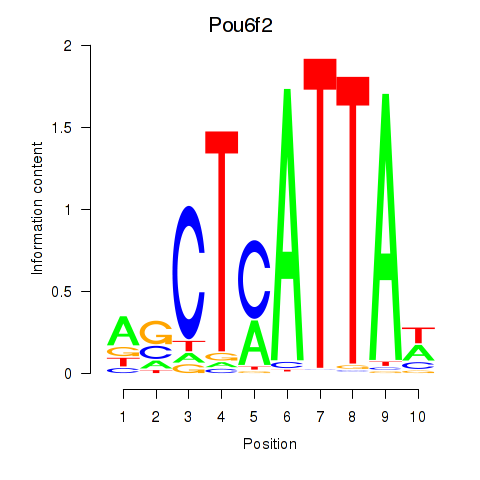
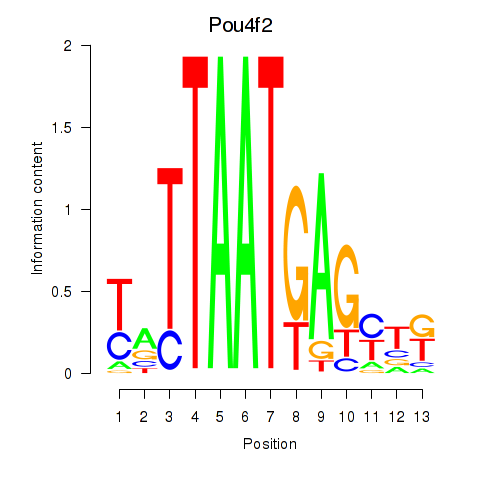
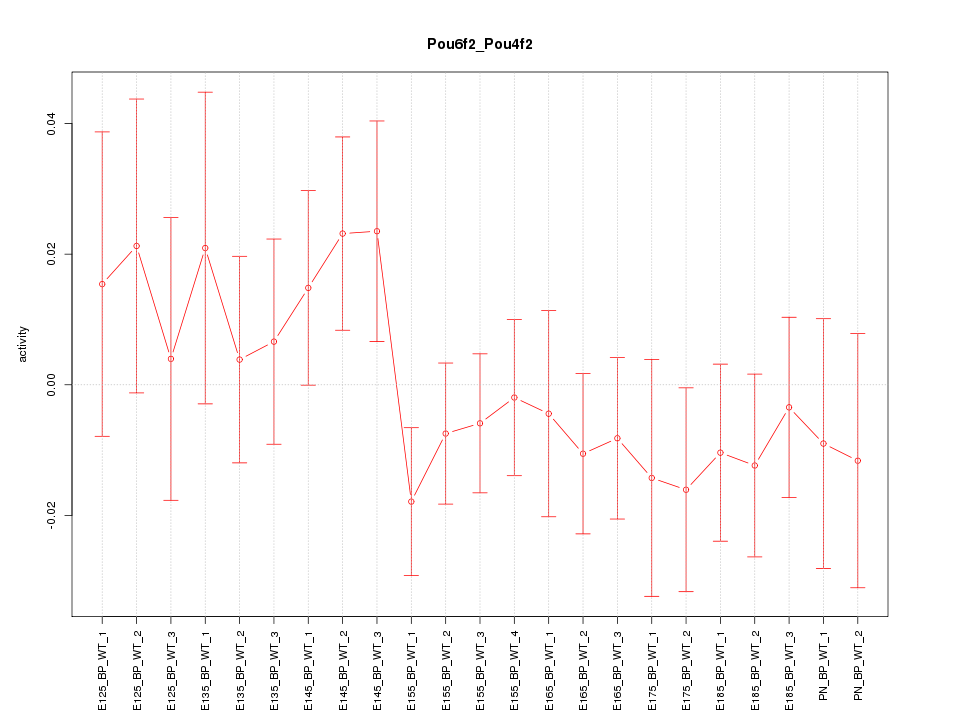
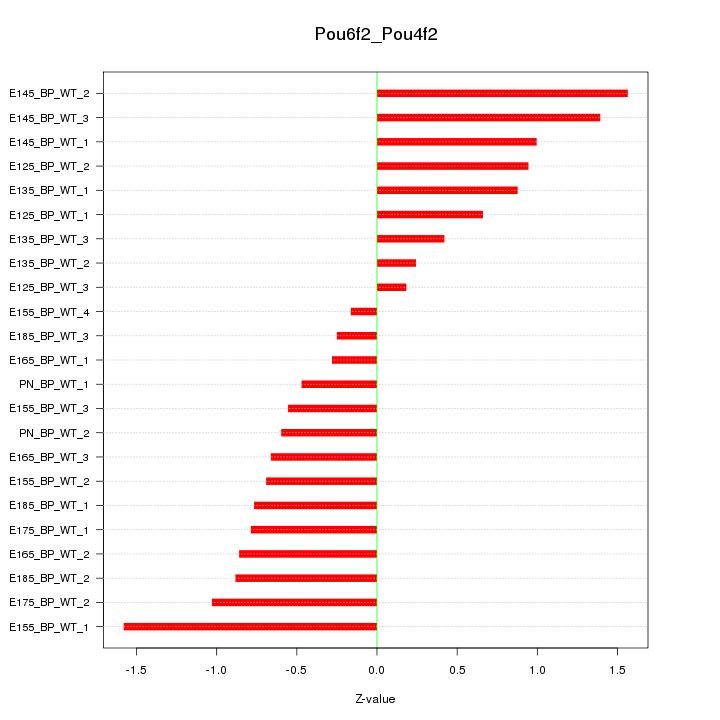
| 2.4 |
7.1 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 1.3 |
16.4 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.9 |
6.8 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 |
3.7 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 |
1.5 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 |
0.9 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.3 |
1.4 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 |
1.3 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.2 |
3.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.2 |
0.5 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 |
0.4 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.1 |
2.7 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
4.5 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
1.2 |
GO:0031424 |
keratinization(GO:0031424) establishment of skin barrier(GO:0061436) |
| 0.1 |
1.0 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 0.1 |
0.6 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
1.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
0.2 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.2 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
0.5 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.4 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.1 |
0.9 |
GO:0048853 |
forebrain morphogenesis(GO:0048853) |
| 0.0 |
2.5 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.2 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.0 |
0.2 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.6 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.1 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 |
0.2 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.0 |
0.2 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) prepulse inhibition(GO:0060134) |
| 0.0 |
0.1 |
GO:0060700 |
regulation of ribonuclease activity(GO:0060700) |
| 0.0 |
0.6 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.6 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.1 |
GO:0032474 |
otolith morphogenesis(GO:0032474) |
| 0.0 |
0.4 |
GO:0097352 |
autophagosome maturation(GO:0097352) |
| 0.0 |
0.1 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 |
0.5 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
0.1 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 |
0.1 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
0.0 |
GO:0032512 |
regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |