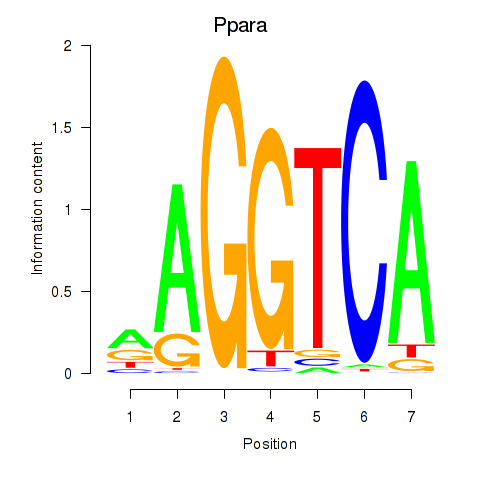
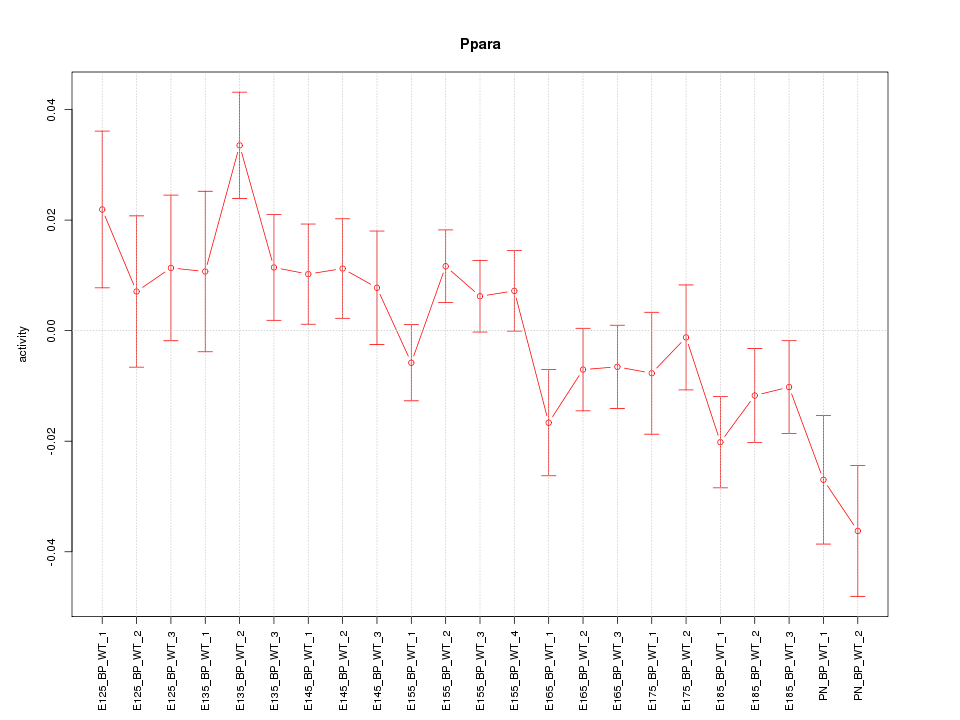
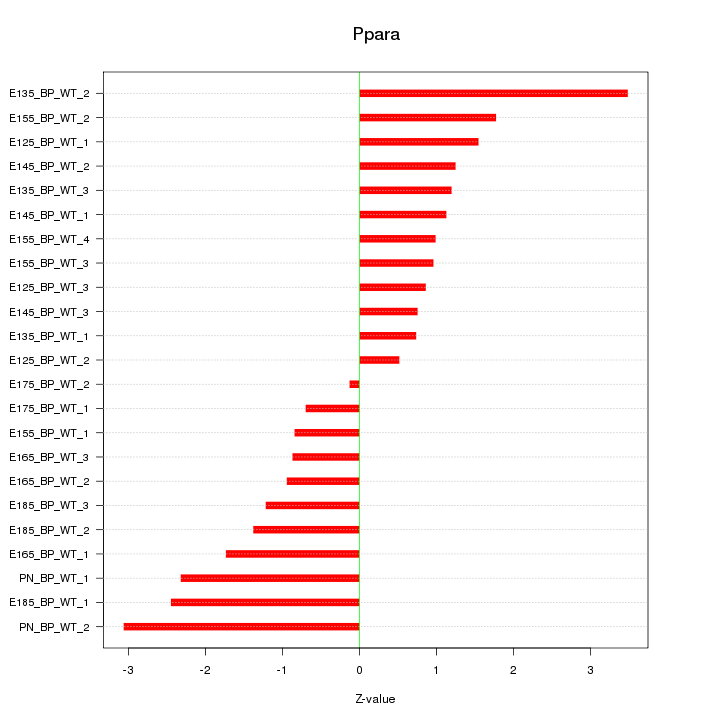
| 1.5 |
9.0 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.3 |
3.8 |
GO:0086017 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) Purkinje myocyte action potential(GO:0086017) membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.2 |
3.6 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 1.0 |
4.1 |
GO:0055095 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.9 |
1.8 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.9 |
2.6 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.9 |
2.6 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.7 |
2.9 |
GO:2000546 |
positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.6 |
3.1 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.6 |
8.3 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.6 |
3.3 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.5 |
2.7 |
GO:0090234 |
cellular response to testosterone stimulus(GO:0071394) regulation of kinetochore assembly(GO:0090234) |
| 0.5 |
3.2 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.5 |
1.6 |
GO:0032493 |
response to bacterial lipoprotein(GO:0032493) |
| 0.5 |
2.1 |
GO:0032275 |
luteinizing hormone secretion(GO:0032275) |
| 0.5 |
5.7 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.5 |
3.1 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.5 |
1.5 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.5 |
3.0 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.5 |
1.5 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.5 |
3.8 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.5 |
1.4 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.5 |
1.4 |
GO:0019405 |
alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.5 |
1.4 |
GO:0072554 |
arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.5 |
1.8 |
GO:0006933 |
negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.5 |
0.9 |
GO:1905065 |
regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.4 |
1.3 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.4 |
1.8 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.4 |
1.3 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 |
4.7 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.4 |
2.2 |
GO:0030240 |
skeletal muscle thin filament assembly(GO:0030240) |
| 0.4 |
1.3 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.4 |
3.8 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.4 |
1.7 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.4 |
2.1 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.4 |
1.2 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.4 |
1.2 |
GO:0042536 |
negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.4 |
1.2 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.4 |
2.0 |
GO:0030951 |
establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 |
1.2 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.4 |
1.1 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.4 |
0.4 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.4 |
1.1 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.4 |
1.8 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 |
1.1 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) hematopoietic stem cell migration(GO:0035701) regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.4 |
3.2 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.4 |
1.8 |
GO:0045794 |
negative regulation of cell volume(GO:0045794) |
| 0.3 |
1.4 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.3 |
1.4 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.3 |
1.0 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 |
1.0 |
GO:0006533 |
aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.3 |
0.7 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.3 |
1.0 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 |
1.3 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.3 |
0.9 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.3 |
1.8 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 |
2.3 |
GO:0038031 |
non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.3 |
0.9 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.3 |
4.3 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.3 |
2.2 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 |
0.8 |
GO:0048259 |
regulation of receptor-mediated endocytosis(GO:0048259) |
| 0.3 |
0.8 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 |
0.8 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 |
1.5 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.3 |
2.8 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 |
0.7 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.2 |
1.9 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.2 |
0.9 |
GO:0010993 |
regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 |
1.2 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
1.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
1.6 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 |
3.2 |
GO:0032211 |
negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.2 |
5.5 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.2 |
3.9 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.2 |
0.9 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.2 |
1.6 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.2 |
0.7 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 |
3.3 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.2 |
0.9 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 |
0.6 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.2 |
1.3 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 |
1.7 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.2 |
0.4 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 |
0.6 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.2 |
0.6 |
GO:0070857 |
negative regulation of receptor biosynthetic process(GO:0010871) regulation of bile acid biosynthetic process(GO:0070857) |
| 0.2 |
2.2 |
GO:0003214 |
cardiac left ventricle morphogenesis(GO:0003214) lung-associated mesenchyme development(GO:0060484) |
| 0.2 |
2.0 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.2 |
1.4 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.2 |
2.6 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.2 |
0.6 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.2 |
8.0 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.2 |
0.6 |
GO:0060261 |
regulation of histone H3-K36 methylation(GO:0000414) positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) |
| 0.2 |
1.4 |
GO:0018342 |
protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.2 |
0.4 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.2 |
0.6 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.2 |
1.7 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.2 |
1.9 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.2 |
0.6 |
GO:0061198 |
fungiform papilla formation(GO:0061198) positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) |
| 0.2 |
2.8 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.2 |
2.3 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 |
3.4 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 |
3.2 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.2 |
2.3 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.2 |
0.3 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.2 |
1.0 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.2 |
0.5 |
GO:0042264 |
peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 |
2.3 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.2 |
5.0 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.2 |
0.6 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.2 |
0.2 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.2 |
1.3 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 |
1.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.2 |
1.6 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 |
0.5 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.2 |
0.5 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.2 |
0.5 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.2 |
0.5 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.1 |
1.0 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
2.1 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.1 |
0.4 |
GO:0021590 |
cerebellum maturation(GO:0021590) cerebellar Purkinje cell layer maturation(GO:0021691) cerebellar cortex maturation(GO:0021699) |
| 0.1 |
0.4 |
GO:0060744 |
thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 |
2.0 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.1 |
0.3 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 |
0.3 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
1.4 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.1 |
1.9 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.1 |
1.9 |
GO:0015937 |
coenzyme A biosynthetic process(GO:0015937) |
| 0.1 |
1.9 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
1.3 |
GO:0097688 |
AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.1 |
3.3 |
GO:1902230 |
negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 |
0.8 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.1 |
0.5 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 |
1.4 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.1 |
2.9 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 |
0.9 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.1 |
1.0 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
5.4 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 |
2.2 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.2 |
GO:2000646 |
positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 |
0.4 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 |
0.8 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 |
0.9 |
GO:0071372 |
cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 |
2.1 |
GO:0048671 |
negative regulation of collateral sprouting(GO:0048671) |
| 0.1 |
1.3 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.1 |
0.2 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.1 |
0.4 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.1 |
3.2 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.1 |
1.1 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
0.2 |
GO:0046533 |
regulation of photoreceptor cell differentiation(GO:0046532) negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.1 |
0.3 |
GO:0050929 |
corticospinal neuron axon guidance(GO:0021966) corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.3 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) tubulin deacetylation(GO:0090042) |
| 0.1 |
0.5 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
0.5 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.1 |
1.6 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.1 |
0.6 |
GO:1903961 |
positive regulation of anion transmembrane transport(GO:1903961) |
| 0.1 |
0.7 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.1 |
0.7 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 |
0.3 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 |
0.5 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.1 |
0.6 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
0.4 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.1 |
2.4 |
GO:0018105 |
peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 |
0.5 |
GO:0000189 |
MAPK import into nucleus(GO:0000189) regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 |
0.3 |
GO:2000612 |
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) metanephric nephron tubule formation(GO:0072289) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 |
2.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.9 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.1 |
0.4 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
0.6 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.1 |
0.3 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.1 |
1.5 |
GO:0014047 |
glutamate secretion(GO:0014047) |
| 0.1 |
0.6 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.1 |
0.5 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 |
0.3 |
GO:0000957 |
mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 |
1.4 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.1 |
0.2 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.5 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.1 |
0.6 |
GO:1903624 |
regulation of apoptotic DNA fragmentation(GO:1902510) regulation of DNA catabolic process(GO:1903624) |
| 0.1 |
0.4 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.6 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.4 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.4 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.2 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.1 |
0.6 |
GO:0032691 |
negative regulation of interleukin-1 beta production(GO:0032691) |
| 0.1 |
1.1 |
GO:0021694 |
cerebellar Purkinje cell layer formation(GO:0021694) |
| 0.1 |
0.6 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.1 |
0.5 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 |
0.3 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.3 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 |
0.5 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 |
0.5 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.5 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.5 |
GO:0061084 |
regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.1 |
0.3 |
GO:1903753 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) hard palate development(GO:0060022) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 |
0.9 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
0.5 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 |
1.8 |
GO:0001782 |
B cell homeostasis(GO:0001782) |
| 0.1 |
0.6 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.2 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.1 |
1.9 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.1 |
0.9 |
GO:0032469 |
endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 |
0.2 |
GO:1903078 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.1 |
2.6 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 |
0.6 |
GO:1900078 |
positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 |
0.7 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
0.5 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 |
0.9 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
2.9 |
GO:0055013 |
cardiac muscle cell development(GO:0055013) |
| 0.1 |
1.9 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
1.1 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
1.8 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.8 |
GO:0014065 |
phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 |
0.2 |
GO:0046103 |
inosine biosynthetic process(GO:0046103) |
| 0.0 |
2.0 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.6 |
GO:0039703 |
viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.7 |
GO:0042491 |
auditory receptor cell differentiation(GO:0042491) |
| 0.0 |
0.3 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
0.1 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.0 |
0.2 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.8 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.7 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.4 |
GO:0032225 |
regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.0 |
1.5 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
4.3 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.0 |
0.5 |
GO:0010259 |
multicellular organism aging(GO:0010259) |
| 0.0 |
0.4 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.5 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.2 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.6 |
GO:0034643 |
establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 |
0.1 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) regulation of potassium ion export(GO:1902302) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 |
0.4 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.4 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.0 |
0.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.2 |
GO:0015692 |
vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.0 |
0.3 |
GO:0051683 |
establishment of Golgi localization(GO:0051683) |
| 0.0 |
0.7 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.5 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.4 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 |
0.1 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.1 |
GO:1904694 |
negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 |
0.1 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 |
0.8 |
GO:0043029 |
T cell homeostasis(GO:0043029) |
| 0.0 |
3.3 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0061386 |
closure of optic fissure(GO:0061386) |
| 0.0 |
0.8 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 |
0.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
2.1 |
GO:0000086 |
G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 |
0.1 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.3 |
GO:0060412 |
ventricular septum morphogenesis(GO:0060412) |
| 0.0 |
1.0 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
3.5 |
GO:0051017 |
actin filament bundle assembly(GO:0051017) |
| 0.0 |
0.2 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.0 |
5.5 |
GO:0043413 |
protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 |
0.2 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
1.7 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.3 |
GO:0002385 |
mucosal immune response(GO:0002385) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.6 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.9 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.7 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
1.2 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.0 |
0.2 |
GO:0031069 |
hair follicle morphogenesis(GO:0031069) |
| 0.0 |
0.9 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.1 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:0061140 |
lung secretory cell differentiation(GO:0061140) |
| 0.0 |
0.5 |
GO:0007224 |
smoothened signaling pathway(GO:0007224) |
| 0.0 |
0.2 |
GO:2000773 |
negative regulation of cellular senescence(GO:2000773) |
| 0.0 |
0.5 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
2.8 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.1 |
GO:0060352 |
cell adhesion molecule production(GO:0060352) |
| 0.0 |
0.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.4 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.2 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.7 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.0 |
GO:0035188 |
blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 |
0.0 |
GO:0048296 |
isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.0 |
0.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.3 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.1 |
GO:0050435 |
beta-amyloid metabolic process(GO:0050435) |
| 0.0 |
0.1 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.2 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.2 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.2 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.4 |
GO:0070830 |
bicellular tight junction assembly(GO:0070830) |
| 0.0 |
0.1 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.0 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.0 |
0.6 |
GO:0006970 |
response to osmotic stress(GO:0006970) |
| 0.0 |
0.1 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0001707 |
mesoderm formation(GO:0001707) mesoderm morphogenesis(GO:0048332) |
| 0.0 |
0.5 |
GO:0010976 |
positive regulation of neuron projection development(GO:0010976) |
| 0.0 |
0.2 |
GO:0045604 |
regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 |
0.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.0 |
0.1 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 |
0.1 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.1 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.0 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.2 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 |
0.1 |
GO:0006366 |
transcription from RNA polymerase II promoter(GO:0006366) |