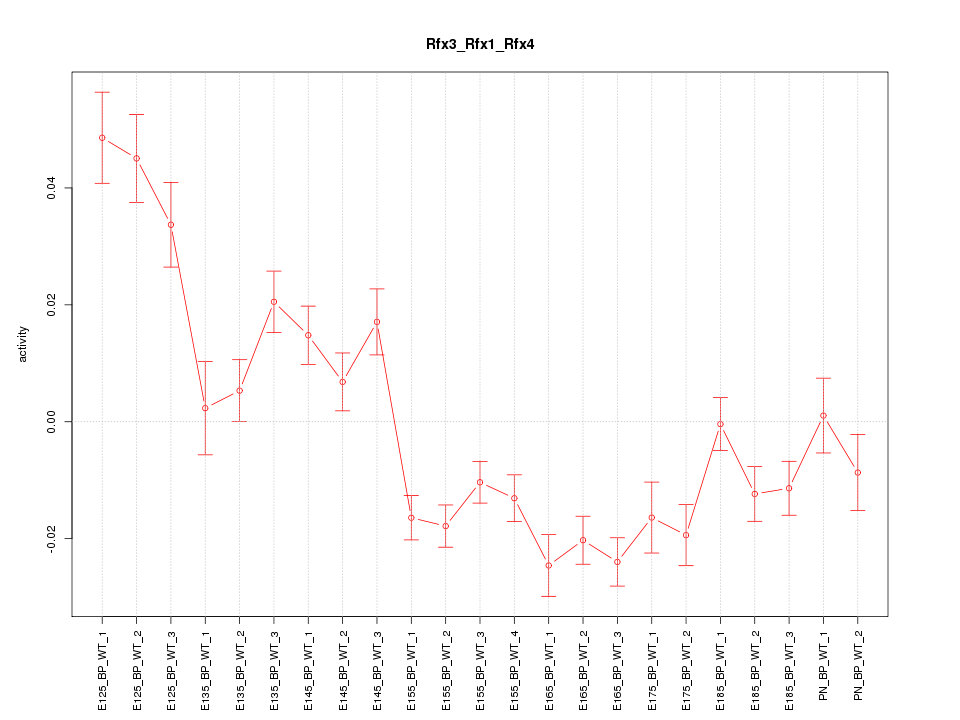
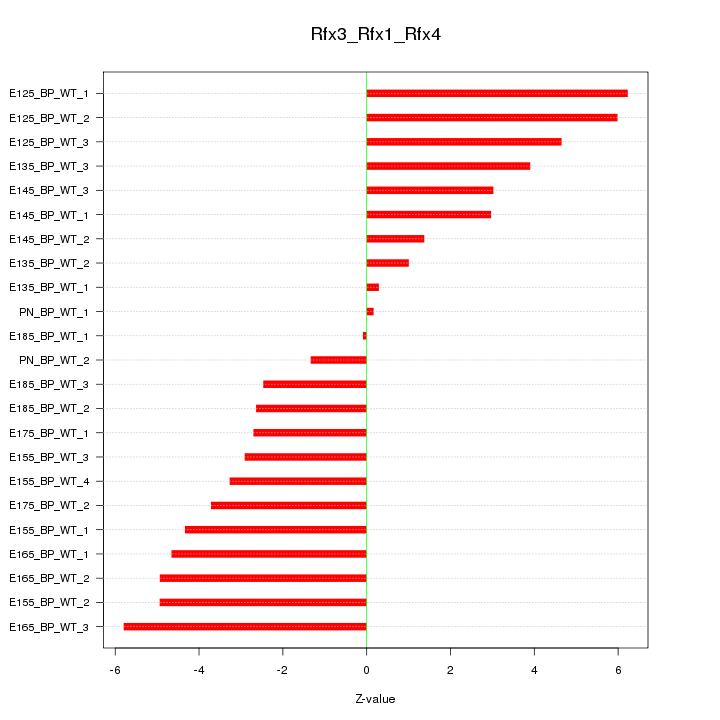
Motif ID: Rfx3_Rfx1_Rfx4
Z-value: 3.675
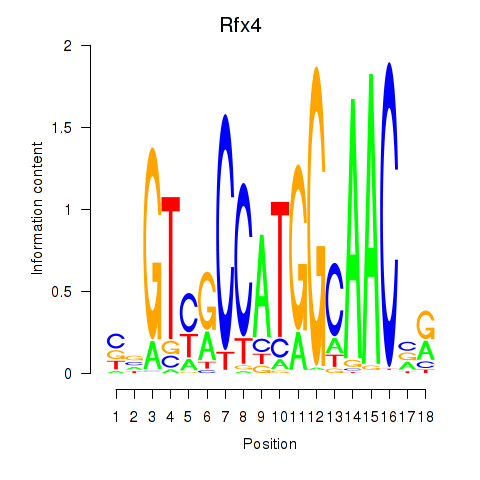
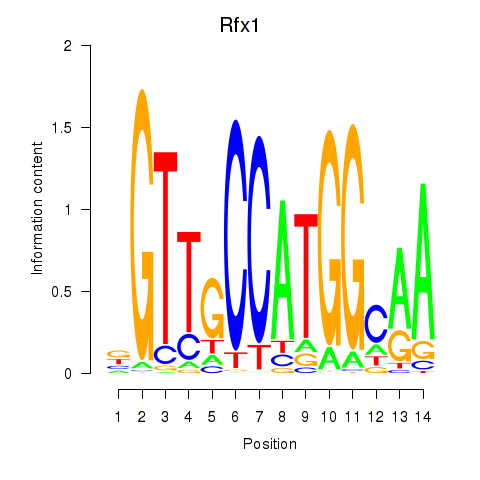
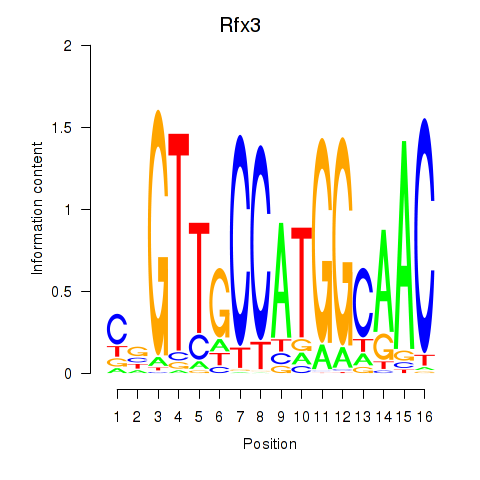
Transcription factors associated with Rfx3_Rfx1_Rfx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rfx1 | ENSMUSG00000031706.6 | Rfx1 |
| Rfx3 | ENSMUSG00000040929.10 | Rfx3 |
| Rfx4 | ENSMUSG00000020037.9 | Rfx4 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx1 | mm10_v2_chr8_+_84066824_84066882 | -0.40 | 5.7e-02 | Click! |
| Rfx4 | mm10_v2_chr10_+_84838143_84838153 | 0.32 | 1.3e-01 | Click! |
| Rfx3 | mm10_v2_chr19_-_28010995_28011054 | -0.04 | 8.5e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 89.7 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 3.2 | 16.0 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 3.0 | 8.9 | GO:0097402 | neuroblast migration(GO:0097402) |
| 2.9 | 11.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 2.8 | 11.3 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 2.0 | 7.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 1.8 | 14.4 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 1.6 | 12.5 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 1.4 | 12.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.4 | 5.6 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 1.3 | 3.8 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 1.2 | 3.6 | GO:2000834 | androgen secretion(GO:0035935) testosterone secretion(GO:0035936) negative regulation of glucagon secretion(GO:0070093) regulation of androgen secretion(GO:2000834) positive regulation of androgen secretion(GO:2000836) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 1.0 | 3.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 1.0 | 21.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 1.0 | 2.9 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 1.0 | 3.8 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.9 | 2.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.9 | 2.6 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.8 | 4.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.7 | 7.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.7 | 4.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.7 | 3.7 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.7 | 2.8 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.7 | 4.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 13.2 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.6 | 1.8 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.6 | 2.9 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.6 | 0.6 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.5 | 6.0 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.5 | 1.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) |
| 0.5 | 2.6 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.5 | 2.0 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.5 | 17.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.5 | 1.9 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.5 | 1.9 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.5 | 2.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.5 | 1.8 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.4 | 5.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 1.3 | GO:1905049 | negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.4 | 2.9 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.4 | 1.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.4 | 2.9 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 | 6.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.3 | 4.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 3.0 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.3 | 22.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.3 | 4.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.3 | 8.1 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.3 | 8.2 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 1.5 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.3 | 1.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.3 | 1.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.3 | 3.5 | GO:0071801 | regulation of podosome assembly(GO:0071801) |
| 0.3 | 0.8 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.3 | 3.6 | GO:0010388 | protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.3 | 1.0 | GO:0090494 | catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.2 | 6.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.0 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.2 | 2.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 3.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 2.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.2 | 0.7 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.2 | 2.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.7 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.2 | 6.8 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 2.7 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.9 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.2 | 0.6 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.2 | 0.6 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 2.9 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.2 | 0.6 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) formin-nucleated actin cable assembly(GO:0070649) |
| 0.2 | 1.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 6.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.2 | 2.4 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.2 | 5.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.2 | 2.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.2 | 1.6 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.2 | 1.3 | GO:0034115 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) cell-cell adhesion involved in gastrulation(GO:0070586) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 1.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 3.1 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.2 | 5.0 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 1.1 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 0.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 1.0 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) paranodal junction assembly(GO:0030913) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 4.7 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.2 | 0.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.2 | 0.8 | GO:0090148 | membrane fission(GO:0090148) |
| 0.2 | 1.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.2 | 1.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 4.0 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 8.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.2 | 1.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 9.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.2 | 0.5 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.1 | 1.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 1.8 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 5.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 13.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.9 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 2.3 | GO:0042517 | positive regulation of tyrosine phosphorylation of Stat3 protein(GO:0042517) |
| 0.1 | 1.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 2.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 1.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 | 0.6 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.1 | 0.6 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 6.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.2 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.3 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 | 0.2 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.1 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.9 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.1 | 0.8 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 3.8 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.4 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 1.8 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.7 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 1.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.1 | 0.2 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.1 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.7 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.1 | 0.3 | GO:0046210 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 1.0 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.6 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 2.8 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.7 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.1 | 0.3 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 1.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.0 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 1.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 0.9 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.7 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 2.3 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 3.0 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 5.1 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 0.2 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 4.1 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.7 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.9 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.6 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 1.1 | GO:0060563 | neuroepithelial cell differentiation(GO:0060563) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 5.1 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.4 | GO:0034766 | negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 | 0.1 | GO:0001193 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 1.0 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.1 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0070613 | regulation of protein processing(GO:0070613) regulation of protein maturation(GO:1903317) |
| 0.0 | 1.2 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 3.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 1.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 1.8 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.7 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 1.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.6 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 2.8 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.1 | GO:0001907 | killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.7 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.2 | GO:0000059 | protein import into nucleus, docking(GO:0000059) |
| 0.0 | 0.4 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.3 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0050965 | sensory perception of temperature stimulus(GO:0050951) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 4.3 | GO:0043269 | regulation of ion transport(GO:0043269) |
| 0.0 | 0.7 | GO:0071426 | ribonucleoprotein complex export from nucleus(GO:0071426) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:1903541 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) exosomal secretion(GO:1990182) |
| 0.0 | 1.0 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 91.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 1.5 | 11.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.8 | 6.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.8 | 3.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.8 | 4.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.7 | 3.7 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.7 | 6.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.6 | 1.8 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.5 | 4.7 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 3.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.4 | 1.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.4 | 4.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 5.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.4 | 4.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.4 | 1.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.3 | 7.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.3 | 5.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.3 | 8.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.3 | 0.8 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 32.0 | GO:0031514 | motile cilium(GO:0031514) |
| 0.2 | 3.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 6.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 4.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.2 | 3.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 5.5 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 1.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 12.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.2 | 0.6 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 2.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 2.7 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.2 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 6.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 2.9 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 3.4 | GO:0001741 | XY body(GO:0001741) |
| 0.2 | 4.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 0.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.1 | 2.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.4 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 4.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 15.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.1 | 3.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.6 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.1 | 16.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 6.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 1.0 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 1.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.1 | 0.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 33.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 3.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 4.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 1.1 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 7.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 2.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.6 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 3.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.5 | GO:0034719 | SMN complex(GO:0032797) SMN-Sm protein complex(GO:0034719) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 2.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 8.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 1.9 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 2.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.3 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 4.7 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 18.4 | GO:0097708 | intracellular vesicle(GO:0097708) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 7.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 10.5 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.5 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.5 | 22.5 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 3.8 | 90.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 3.2 | 19.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 2.6 | 12.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 2.3 | 6.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 1.4 | 5.7 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 1.4 | 4.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.1 | 6.8 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 1.1 | 10.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 1.0 | 4.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 1.0 | 7.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 1.0 | 2.9 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 1.0 | 2.9 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.9 | 4.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.8 | 3.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.7 | 6.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.7 | 2.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.7 | 16.0 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.7 | 2.7 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.5 | 5.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.5 | 2.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.5 | 1.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 8.9 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.5 | 1.9 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.4 | 0.4 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.3 | 1.0 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.3 | 1.7 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.3 | 2.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.3 | 7.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 0.6 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.3 | 0.8 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.3 | 1.3 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 1.0 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.3 | 9.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 7.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 1.9 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.2 | 1.6 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 1.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.2 | 2.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.7 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 3.4 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.2 | 3.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.2 | 14.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 13.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 0.6 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) U7 snRNA binding(GO:0071209) |
| 0.2 | 1.0 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.2 | 2.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 2.0 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 4.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.2 | 2.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.2 | 1.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 6.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 6.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 2.8 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 9.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 6.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 17.9 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 3.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 3.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 5.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 5.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.0 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.0 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 3.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.6 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.4 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.9 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.4 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 3.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.5 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 4.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 1.6 | GO:0045502 | dynein binding(GO:0045502) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.3 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.4 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.1 | 4.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 1.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 2.0 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 2.8 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 2.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 2.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 1.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 5.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0008905 | mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.6 | GO:0034945 | dihydrolipoamide branched chain acyltransferase activity(GO:0004147) palmitoleoyl [acyl-carrier-protein]-dependent acyltransferase activity(GO:0008951) serine O-acyltransferase activity(GO:0016412) O-succinyltransferase activity(GO:0016750) sinapoyltransferase activity(GO:0016752) O-sinapoyltransferase activity(GO:0016753) peptidyl-lysine N6-myristoyltransferase activity(GO:0018030) peptidyl-lysine N6-palmitoyltransferase activity(GO:0018031) benzoyl acetate-CoA thiolase activity(GO:0018711) 3-hydroxybutyryl-CoA thiolase activity(GO:0018712) 3-ketopimelyl-CoA thiolase activity(GO:0018713) N-palmitoyltransferase activity(GO:0019105) acyl-CoA N-acyltransferase activity(GO:0019186) protein-cysteine S-myristoyltransferase activity(GO:0019705) glucosaminyl-phosphotidylinositol O-acyltransferase activity(GO:0032216) ergosterol O-acyltransferase activity(GO:0034737) lanosterol O-acyltransferase activity(GO:0034738) naphthyl-2-oxomethyl-succinyl-CoA succinyl transferase activity(GO:0034848) 2,4,4-trimethyl-3-oxopentanoyl-CoA 2-C-propanoyl transferase activity(GO:0034851) 2-methylhexanoyl-CoA C-acetyltransferase activity(GO:0034915) butyryl-CoA 2-C-propionyltransferase activity(GO:0034919) 2,6-dimethyl-5-methylene-3-oxo-heptanoyl-CoA C-acetyltransferase activity(GO:0034945) L-2-aminoadipate N-acetyltransferase activity(GO:0043741) keto acid formate lyase activity(GO:0043806) azetidine-2-carboxylic acid acetyltransferase activity(GO:0046941) peptidyl-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0052858) acetyl-CoA:L-lysine N6-acetyltransferase(GO:0090595) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) |
| 0.0 | 0.5 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.5 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 10.1 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |