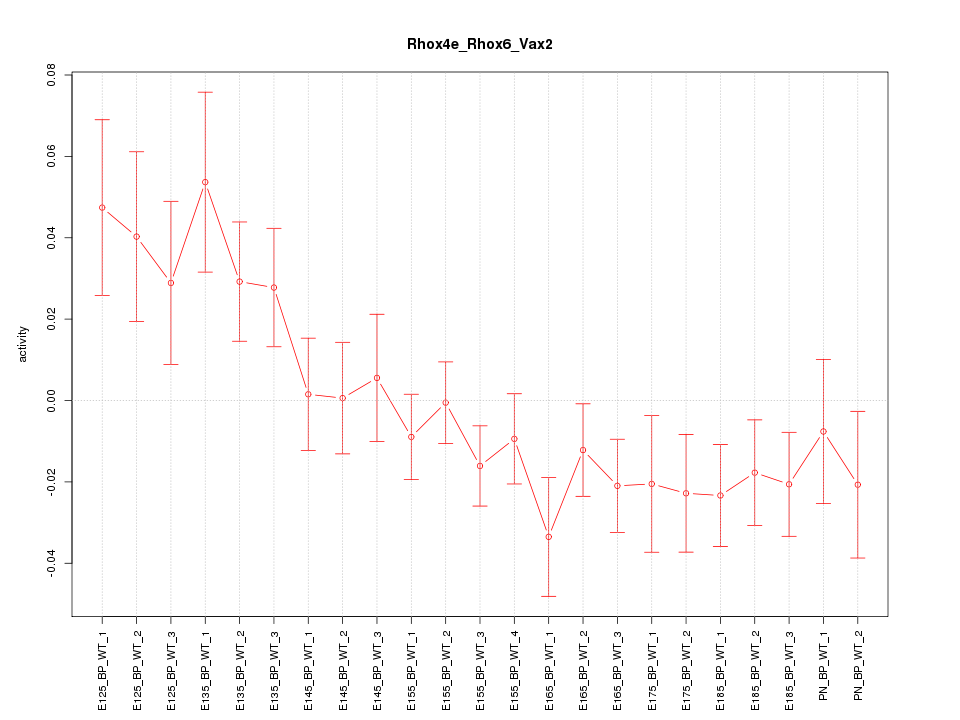
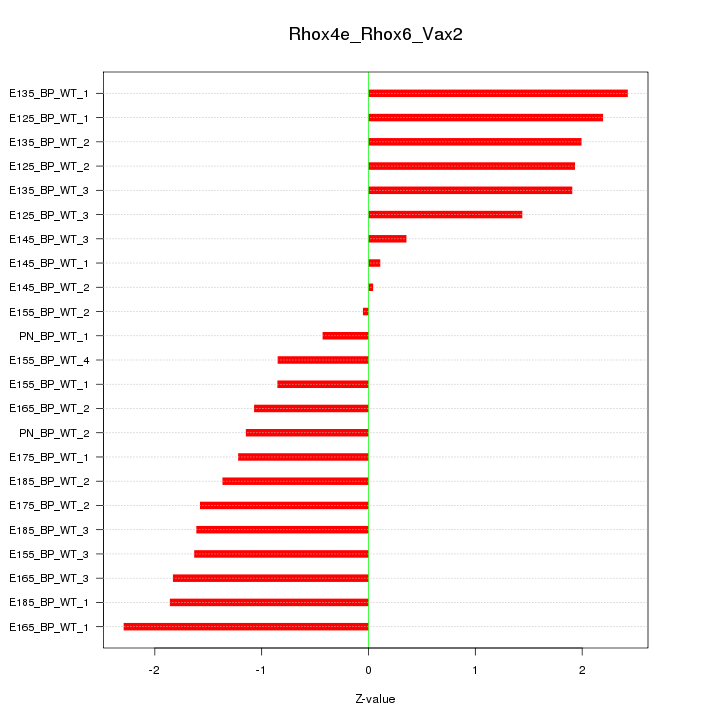
Motif ID: Rhox4e_Rhox6_Vax2
Z-value: 1.497
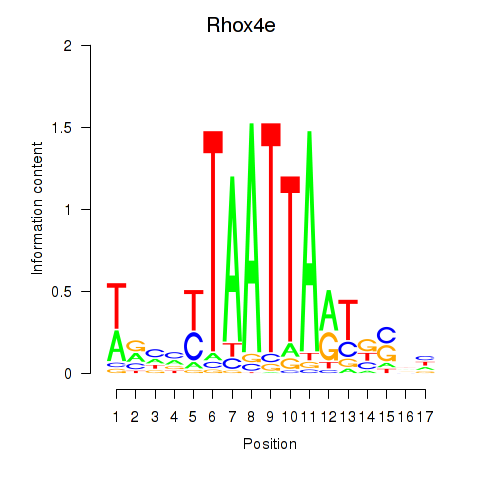
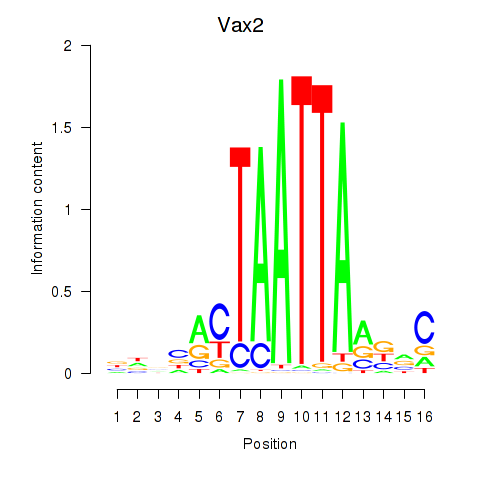
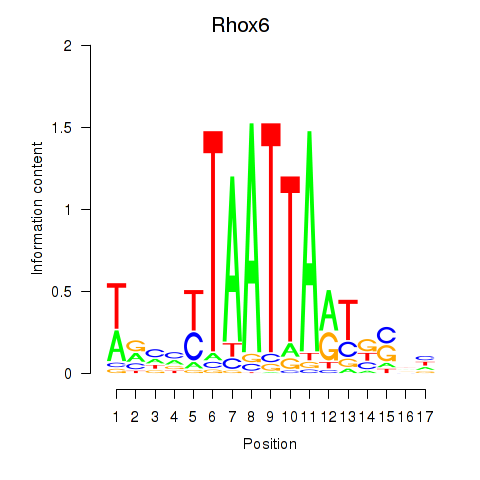
Transcription factors associated with Rhox4e_Rhox6_Vax2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Rhox4e | ENSMUSG00000071770.4 | Rhox4e |
| Rhox6 | ENSMUSG00000006200.3 | Rhox6 |
| Vax2 | ENSMUSG00000034777.1 | Vax2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vax2 | mm10_v2_chr6_+_83711232_83711288 | -0.17 | 4.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 26.9 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.9 | 2.8 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.8 | 5.5 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.6 | 10.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.5 | 2.6 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.5 | 16.5 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.4 | 4.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) |
| 0.4 | 0.7 | GO:0061738 | late endosomal microautophagy(GO:0061738) |
| 0.3 | 3.1 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 1.0 | GO:0048818 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.3 | 0.9 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.3 | 5.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.3 | 1.3 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.2 | 1.0 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.2 | 5.0 | GO:0007614 | short-term memory(GO:0007614) |
| 0.2 | 4.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.2 | 0.6 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.2 | 0.6 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.2 | 0.7 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.2 | 0.7 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.0 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 1.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.2 | 2.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 3.6 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.1 | 3.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.9 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 | 0.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 3.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 3.1 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.1 | 0.4 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 2.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.7 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.5 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 3.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 1.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.2 | GO:0015819 | lysine transport(GO:0015819) |
| 0.0 | 0.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 2.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 10.4 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.5 | GO:0006337 | nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.4 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 2.9 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.8 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.4 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.0 | 0.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 1.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.2 | GO:0051900 | regulation of mitochondrial depolarization(GO:0051900) |
| 0.0 | 2.0 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.7 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.1 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) dendritic spine neck(GO:0044326) |
| 0.4 | 1.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 1.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 2.6 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.8 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 2.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 2.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 3.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 2.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 4.9 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.6 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 2.8 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 0.7 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.0 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 5.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 17.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 4.1 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 2.2 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.1 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0030689 | preribosome, small subunit precursor(GO:0030688) Noc complex(GO:0030689) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.5 | 4.3 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.5 | 3.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 10.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 1.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 1.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 27.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 1.0 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.7 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 5.5 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 2.8 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 1.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 2.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 4.8 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.1 | 0.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.4 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 9.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.7 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0052740 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.4 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 2.2 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 1.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 1.1 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 1.0 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.9 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 1.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 2.0 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 3.5 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 11.6 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0008198 | ATPase activator activity(GO:0001671) ferrous iron binding(GO:0008198) |
| 0.0 | 0.7 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 4.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 7.8 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |