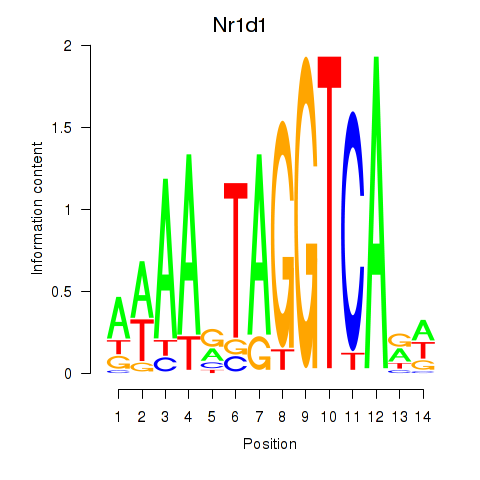
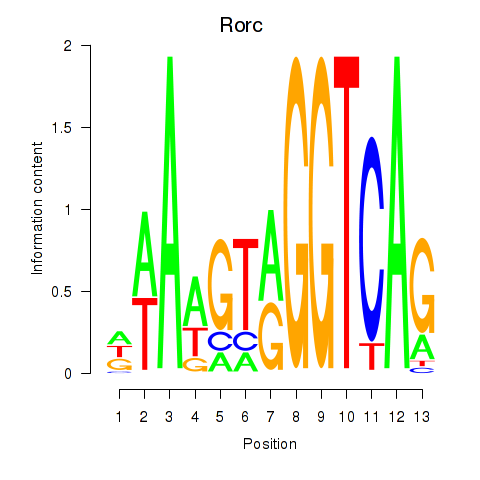
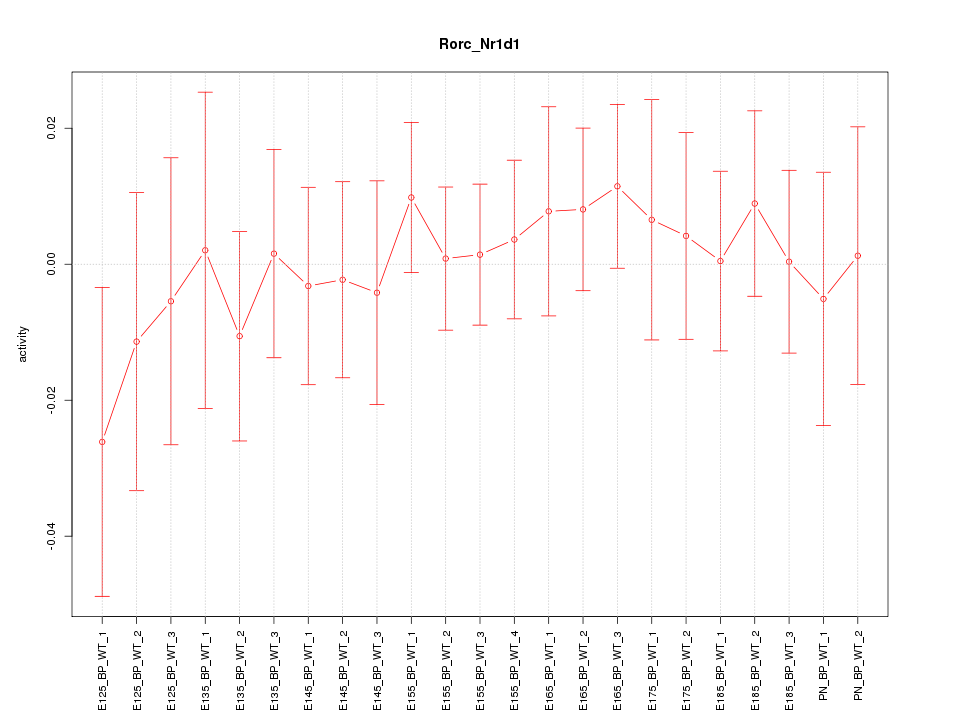
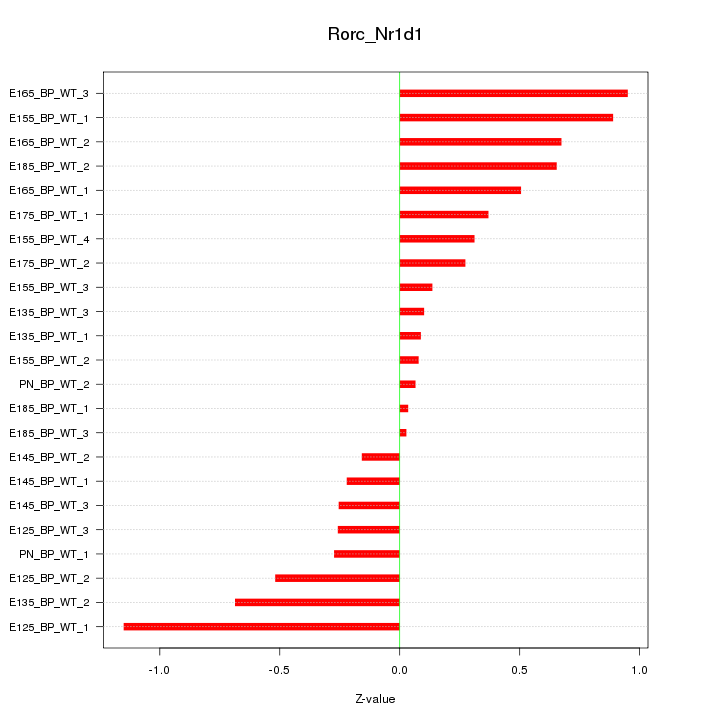
| 0.4 |
1.3 |
GO:0090403 |
oxidative stress-induced premature senescence(GO:0090403) |
| 0.2 |
1.3 |
GO:0072513 |
positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 |
0.5 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.2 |
0.5 |
GO:0060084 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.2 |
1.6 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 |
1.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.3 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
0.1 |
GO:0007113 |
endomitotic cell cycle(GO:0007113) |
| 0.1 |
0.2 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 |
0.1 |
GO:0050859 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) negative regulation of serotonin secretion(GO:0014063) negative regulation of B cell receptor signaling pathway(GO:0050859) regulation of eosinophil activation(GO:1902566) negative regulation of membrane invagination(GO:1905154) |
| 0.0 |
0.1 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.0 |
0.1 |
GO:0031627 |
telomeric loop formation(GO:0031627) |
| 0.0 |
0.6 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.0 |
0.8 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
2.6 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0014053 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 |
0.0 |
GO:1904995 |
negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 |
1.0 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.0 |
0.2 |
GO:0043206 |
extracellular fibril organization(GO:0043206) |
| 0.0 |
0.3 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 |
0.1 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.0 |
0.3 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.0 |
0.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.1 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.0 |
0.0 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.0 |
0.2 |
GO:0033572 |
transferrin transport(GO:0033572) |
| 0.0 |
0.4 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.0 |
GO:0002741 |
positive regulation of cytokine secretion involved in immune response(GO:0002741) |