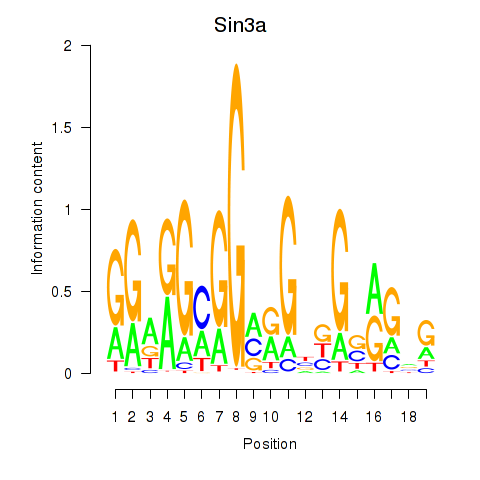
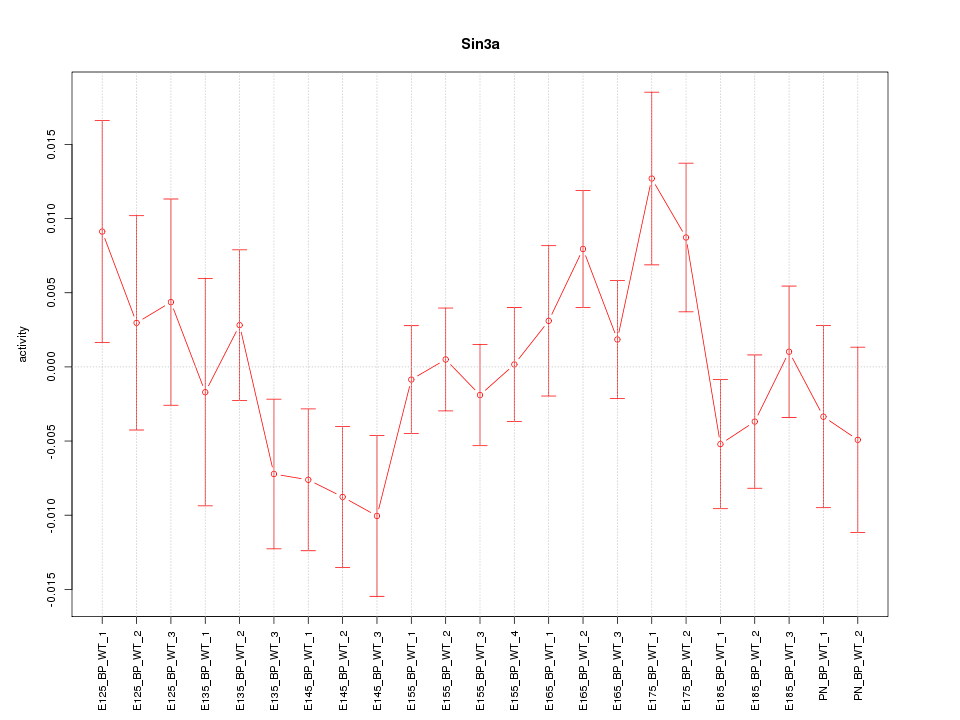
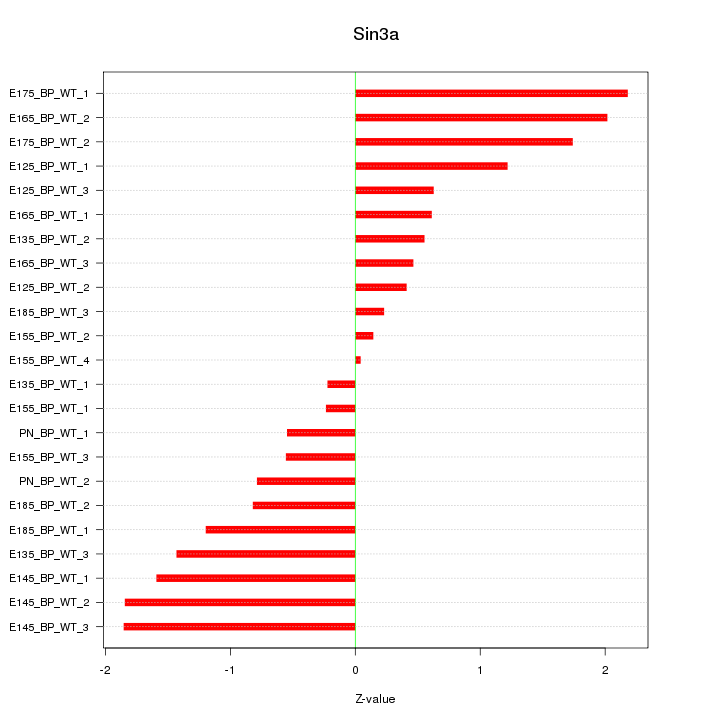
| 0.7 |
0.7 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.7 |
4.3 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.7 |
3.3 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.5 |
1.5 |
GO:0016191 |
synaptic vesicle uncoating(GO:0016191) regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.5 |
4.0 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.5 |
1.5 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.5 |
1.4 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.4 |
1.3 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 |
1.3 |
GO:0035604 |
fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.4 |
1.2 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.4 |
1.2 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.4 |
1.2 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.4 |
1.6 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.4 |
2.9 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.4 |
1.5 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.4 |
2.5 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.4 |
1.4 |
GO:0051643 |
endoplasmic reticulum localization(GO:0051643) |
| 0.4 |
1.1 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.4 |
1.8 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 |
1.7 |
GO:0003404 |
optic vesicle morphogenesis(GO:0003404) |
| 0.3 |
1.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 |
1.6 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.3 |
1.3 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.3 |
1.0 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.3 |
1.5 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.3 |
1.2 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.3 |
1.1 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.3 |
0.9 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 |
2.5 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 |
1.7 |
GO:0035469 |
determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.3 |
1.1 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 |
1.0 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.3 |
1.3 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
2.0 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 |
0.2 |
GO:0071910 |
determination of liver left/right asymmetry(GO:0071910) |
| 0.2 |
0.9 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.2 |
0.9 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
0.9 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.2 |
1.4 |
GO:0070863 |
positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 |
1.6 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.2 |
1.3 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 |
0.7 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.2 |
1.1 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 |
2.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.2 |
0.4 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.2 |
0.6 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 |
3.7 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.2 |
1.4 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 |
0.6 |
GO:0060364 |
frontal suture morphogenesis(GO:0060364) |
| 0.2 |
0.6 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 |
0.6 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 |
0.6 |
GO:0007290 |
spermatid nucleus elongation(GO:0007290) |
| 0.2 |
0.9 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.2 |
0.4 |
GO:0050856 |
regulation of T cell receptor signaling pathway(GO:0050856) |
| 0.2 |
0.6 |
GO:0048352 |
neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 0.2 |
0.6 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.2 |
1.1 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 |
0.7 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 |
0.5 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 |
0.4 |
GO:0032418 |
lysosome localization(GO:0032418) |
| 0.2 |
1.4 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 |
0.9 |
GO:0097107 |
postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) |
| 0.2 |
0.2 |
GO:0071335 |
hair follicle cell proliferation(GO:0071335) |
| 0.2 |
1.0 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.2 |
1.9 |
GO:0046548 |
retinal rod cell development(GO:0046548) |
| 0.2 |
1.9 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.2 |
0.5 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.2 |
1.0 |
GO:0044829 |
positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 |
1.2 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.2 |
0.3 |
GO:1902263 |
apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.2 |
1.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 |
0.7 |
GO:0000255 |
allantoin metabolic process(GO:0000255) |
| 0.2 |
1.2 |
GO:0035372 |
protein localization to microtubule(GO:0035372) |
| 0.2 |
0.3 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 |
0.3 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.2 |
0.5 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
1.0 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.2 |
2.0 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.2 |
1.4 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.2 |
0.3 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.2 |
1.1 |
GO:1904017 |
positive regulation of female receptivity(GO:0045925) response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 |
1.2 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 |
0.8 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 |
0.6 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 |
2.9 |
GO:0031442 |
positive regulation of mRNA 3'-end processing(GO:0031442) |
| 0.2 |
0.8 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.2 |
0.6 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.4 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.1 |
0.4 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.1 |
1.6 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
0.4 |
GO:0006065 |
UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.1 |
0.4 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.1 |
1.4 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
1.0 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.4 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
1.5 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.1 |
0.3 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 |
0.3 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.1 |
0.4 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 |
0.5 |
GO:0010701 |
positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 |
1.0 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.1 |
GO:0060994 |
regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.1 |
0.6 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 |
0.5 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.1 |
0.4 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 |
0.5 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.8 |
GO:0000056 |
ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 |
0.4 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.1 |
0.4 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 |
0.5 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.1 |
0.4 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.1 |
0.7 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.6 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
1.0 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.1 |
0.2 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.2 |
GO:2001012 |
mesenchymal cell differentiation involved in kidney development(GO:0072161) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.1 |
0.2 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.1 |
0.1 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 0.1 |
0.3 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.3 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.1 |
0.6 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
0.7 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
0.3 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
0.5 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.2 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.1 |
0.9 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 |
0.5 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
1.6 |
GO:0045722 |
positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 |
0.3 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.5 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.1 |
1.1 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.4 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 0.1 |
0.4 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.1 |
0.3 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.1 |
0.6 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.1 |
GO:0072368 |
regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.1 |
1.0 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.4 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 |
2.1 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.4 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.1 |
0.3 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.3 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
0.5 |
GO:0070305 |
response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 0.1 |
0.4 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 |
0.4 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 |
0.5 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.1 |
0.4 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.2 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
0.7 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.1 |
0.5 |
GO:0003344 |
pericardium morphogenesis(GO:0003344) |
| 0.1 |
0.2 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 |
1.0 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) |
| 0.1 |
0.9 |
GO:0032252 |
secretory granule localization(GO:0032252) |
| 0.1 |
0.2 |
GO:0003192 |
mitral valve formation(GO:0003192) |
| 0.1 |
0.6 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 |
0.3 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 |
0.5 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
0.9 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
0.7 |
GO:0048570 |
notochord morphogenesis(GO:0048570) |
| 0.1 |
0.8 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.5 |
GO:0032229 |
negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.1 |
0.9 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
1.1 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.3 |
GO:0065001 |
negative regulation of histone H3-K36 methylation(GO:0000415) specification of axis polarity(GO:0065001) |
| 0.1 |
0.6 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 |
0.3 |
GO:0030050 |
vesicle transport along actin filament(GO:0030050) |
| 0.1 |
0.9 |
GO:0003283 |
atrial septum development(GO:0003283) |
| 0.1 |
0.2 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) |
| 0.1 |
0.8 |
GO:0042492 |
gamma-delta T cell differentiation(GO:0042492) |
| 0.1 |
0.2 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 |
0.7 |
GO:2000794 |
regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.1 |
0.6 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.2 |
GO:0051695 |
actin filament uncapping(GO:0051695) |
| 0.1 |
0.5 |
GO:0098543 |
detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 |
0.7 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 |
0.4 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.6 |
GO:0071044 |
histone mRNA catabolic process(GO:0071044) |
| 0.1 |
0.7 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 |
0.3 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.1 |
GO:0014063 |
negative regulation of serotonin secretion(GO:0014063) |
| 0.1 |
0.9 |
GO:0030948 |
negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 |
0.7 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 |
0.4 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 |
1.1 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.1 |
0.1 |
GO:1902566 |
regulation of eosinophil activation(GO:1902566) |
| 0.1 |
0.3 |
GO:0055099 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) regulation of cholesterol esterification(GO:0010872) response to high density lipoprotein particle(GO:0055099) |
| 0.1 |
0.4 |
GO:0010747 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 |
0.1 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.6 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
1.3 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 |
0.6 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 0.1 |
0.6 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 |
0.3 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.3 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
0.1 |
GO:1902916 |
positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 |
0.2 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction(GO:0014819) |
| 0.1 |
0.3 |
GO:0034145 |
positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.1 |
0.5 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 |
0.1 |
GO:0060594 |
mammary gland specification(GO:0060594) |
| 0.1 |
0.5 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.7 |
GO:0036120 |
cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 |
0.7 |
GO:1904152 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) regulation of retrograde protein transport, ER to cytosol(GO:1904152) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
0.2 |
GO:0050862 |
positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 |
0.2 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.1 |
0.1 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 |
0.4 |
GO:0060763 |
mammary duct terminal end bud growth(GO:0060763) |
| 0.1 |
0.2 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
1.3 |
GO:0010172 |
embryonic body morphogenesis(GO:0010172) |
| 0.1 |
0.5 |
GO:0008354 |
germ cell migration(GO:0008354) |
| 0.1 |
0.2 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 |
0.3 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.7 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.1 |
0.8 |
GO:0055090 |
acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.1 |
0.3 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
0.2 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 |
1.0 |
GO:0045653 |
negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.1 |
0.1 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.9 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.1 |
0.3 |
GO:0036089 |
cleavage furrow formation(GO:0036089) |
| 0.1 |
0.2 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.1 |
0.2 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 |
0.7 |
GO:0086012 |
membrane depolarization during cardiac muscle cell action potential(GO:0086012) |
| 0.1 |
1.0 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.1 |
0.4 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.1 |
0.1 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 0.1 |
0.6 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
0.5 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.2 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.7 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
1.1 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.7 |
GO:0050910 |
detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 |
0.6 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.3 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.1 |
0.2 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 |
0.2 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 |
0.5 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.1 |
0.7 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.1 |
GO:0010730 |
negative regulation of hydrogen peroxide metabolic process(GO:0010727) negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 |
0.3 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.9 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.1 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.4 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
0.3 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.1 |
2.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
0.2 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.2 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.1 |
0.2 |
GO:2000049 |
positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.1 |
0.2 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.1 |
0.1 |
GO:0070734 |
histone H3-K27 methylation(GO:0070734) |
| 0.1 |
0.4 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.1 |
0.3 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
0.4 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 |
0.2 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.1 |
0.2 |
GO:2000052 |
positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 |
0.4 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.1 |
0.2 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.2 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.2 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.0 |
1.1 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.2 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 0.0 |
0.1 |
GO:0030421 |
defecation(GO:0030421) |
| 0.0 |
0.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.0 |
0.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.0 |
0.1 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 |
0.2 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 |
0.1 |
GO:0060907 |
positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 |
0.4 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.1 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.0 |
0.1 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.2 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.0 |
0.5 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.0 |
0.2 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.4 |
GO:1901142 |
insulin metabolic process(GO:1901142) |
| 0.0 |
0.6 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.3 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
0.2 |
GO:2000587 |
regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.3 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.7 |
GO:1901224 |
positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 |
0.2 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 0.0 |
0.7 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.7 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 |
0.3 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.0 |
0.2 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.0 |
0.2 |
GO:0034397 |
telomere localization(GO:0034397) |
| 0.0 |
0.7 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) |
| 0.0 |
0.2 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.1 |
GO:0032509 |
endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 |
0.0 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 |
0.1 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) female courtship behavior(GO:0008050) regulation of female receptivity(GO:0045924) |
| 0.0 |
0.1 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 |
0.2 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.0 |
1.2 |
GO:0021952 |
central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 |
0.2 |
GO:0010825 |
positive regulation of centrosome duplication(GO:0010825) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.4 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.6 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.3 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.0 |
0.5 |
GO:0043923 |
positive regulation by host of viral transcription(GO:0043923) |
| 0.0 |
0.2 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.3 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:1903519 |
regulation of acrosome reaction(GO:0060046) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 |
0.2 |
GO:0070537 |
histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 |
0.1 |
GO:0010920 |
negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.2 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
1.1 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 |
0.1 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 |
0.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.0 |
0.1 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.1 |
GO:0006030 |
chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 |
0.3 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.2 |
GO:0071415 |
cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 |
0.2 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 |
1.0 |
GO:0097009 |
energy homeostasis(GO:0097009) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.2 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
1.0 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.3 |
GO:0060052 |
neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 |
0.1 |
GO:0006083 |
acetate metabolic process(GO:0006083) |
| 0.0 |
0.2 |
GO:0019321 |
pentose metabolic process(GO:0019321) |
| 0.0 |
0.2 |
GO:0018125 |
peptidyl-cysteine methylation(GO:0018125) |
| 0.0 |
0.4 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.1 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.1 |
GO:2000664 |
positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 |
0.2 |
GO:0051697 |
protein delipidation(GO:0051697) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.0 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.3 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 |
0.3 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 |
0.3 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
1.0 |
GO:0007492 |
endoderm development(GO:0007492) |
| 0.0 |
0.5 |
GO:0032608 |
interferon-beta production(GO:0032608) |
| 0.0 |
0.1 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 |
0.4 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.0 |
0.6 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.5 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
0.3 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.0 |
0.1 |
GO:0001869 |
regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.0 |
0.1 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 |
0.7 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.5 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.7 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.4 |
GO:0032800 |
receptor biosynthetic process(GO:0032800) |
| 0.0 |
0.2 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.0 |
0.2 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.0 |
0.2 |
GO:0086018 |
T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) SA node cell action potential(GO:0086015) SA node cell to atrial cardiac muscle cell signalling(GO:0086018) SA node cell to atrial cardiac muscle cell communication(GO:0086070) regulation of SA node cell action potential(GO:0098907) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.2 |
GO:0035561 |
regulation of chromatin binding(GO:0035561) |
| 0.0 |
0.1 |
GO:2000110 |
protein sialylation(GO:1990743) negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 |
0.1 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.5 |
GO:0030851 |
granulocyte differentiation(GO:0030851) |
| 0.0 |
0.7 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.0 |
0.4 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.0 |
0.2 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.3 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.6 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.0 |
0.1 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 0.0 |
0.2 |
GO:0010603 |
regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 |
0.4 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.0 |
0.2 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.0 |
0.2 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.0 |
GO:0060295 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 |
0.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.5 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 |
0.0 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 |
0.1 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.1 |
GO:0040014 |
regulation of multicellular organism growth(GO:0040014) |
| 0.0 |
0.1 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.0 |
0.0 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.0 |
0.3 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |
| 0.0 |
0.2 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.2 |
GO:0042438 |
melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.2 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.0 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.7 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.0 |
0.1 |
GO:0001973 |
adenosine receptor signaling pathway(GO:0001973) |
| 0.0 |
0.4 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.2 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.4 |
GO:0007569 |
cell aging(GO:0007569) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.2 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.4 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.2 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.2 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.0 |
0.4 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.0 |
0.3 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.2 |
GO:0034377 |
plasma lipoprotein particle assembly(GO:0034377) |
| 0.0 |
0.3 |
GO:0043330 |
response to exogenous dsRNA(GO:0043330) |
| 0.0 |
0.2 |
GO:0046499 |
S-adenosylmethioninamine metabolic process(GO:0046499) |
| 0.0 |
0.3 |
GO:0001960 |
negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 |
0.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.1 |
GO:0032484 |
Ral protein signal transduction(GO:0032484) |
| 0.0 |
0.1 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.0 |
0.1 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.5 |
GO:0006493 |
protein O-linked glycosylation(GO:0006493) |
| 0.0 |
0.0 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.1 |
GO:0051883 |
disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 |
0.2 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.1 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.1 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.0 |
0.0 |
GO:2000054 |
regulation of centromeric sister chromatid cohesion(GO:0070602) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 |
0.1 |
GO:0035413 |
positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 |
0.1 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.0 |
GO:0042447 |
hormone catabolic process(GO:0042447) |
| 0.0 |
0.2 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.0 |
0.1 |
GO:0086001 |
cardiac muscle cell action potential(GO:0086001) |
| 0.0 |
0.0 |
GO:0031622 |
fever generation(GO:0001660) regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) regulation of penile erection(GO:0060405) |
| 0.0 |
0.2 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.2 |
GO:0006308 |
DNA catabolic process(GO:0006308) |
| 0.0 |
0.2 |
GO:0021516 |
dorsal spinal cord development(GO:0021516) |
| 0.0 |
0.3 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 |
0.1 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.0 |
0.1 |
GO:0036289 |
peptidyl-serine autophosphorylation(GO:0036289) |
| 0.0 |
0.6 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.1 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.0 |
1.1 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.1 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.1 |
GO:0097084 |
vascular smooth muscle cell development(GO:0097084) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.1 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.1 |
GO:0035066 |
positive regulation of histone acetylation(GO:0035066) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.1 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.2 |
GO:0007143 |
female meiotic division(GO:0007143) |
| 0.0 |
0.5 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
1.0 |
GO:0032200 |
telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 |
0.1 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.1 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0051125 |
regulation of actin nucleation(GO:0051125) |
| 0.0 |
0.0 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:0006706 |
steroid catabolic process(GO:0006706) cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.5 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.0 |
GO:0046210 |
nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 |
0.0 |
GO:0045875 |
negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 |
0.1 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.1 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.1 |
GO:0003009 |
skeletal muscle contraction(GO:0003009) |
| 0.0 |
0.2 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.3 |
GO:0010390 |
histone monoubiquitination(GO:0010390) |