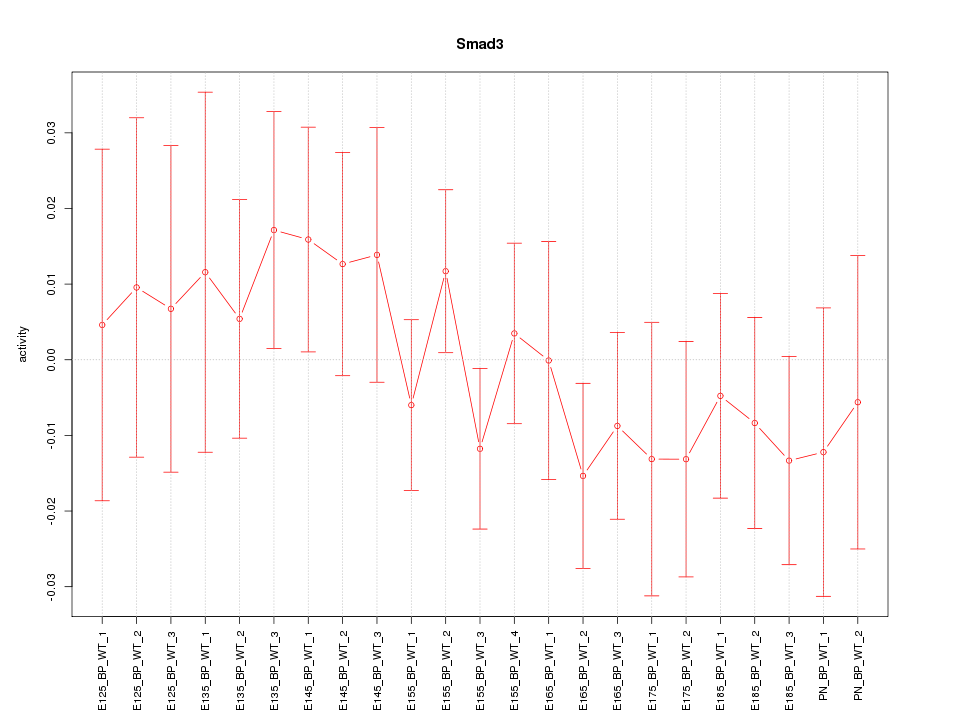
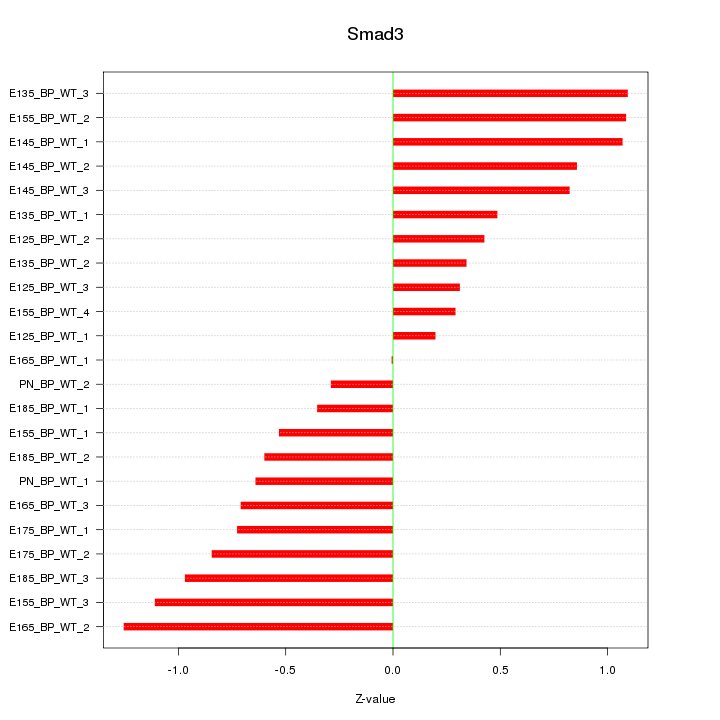
Motif ID: Smad3
Z-value: 0.736
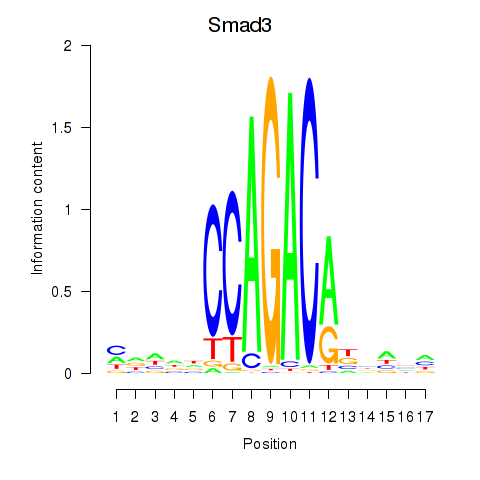
Transcription factors associated with Smad3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Smad3 | ENSMUSG00000032402.6 | Smad3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad3 | mm10_v2_chr9_-_63757933_63757994 | 0.66 | 6.0e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.6 | 1.7 | GO:1905065 | hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.3 | 0.9 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.3 | 5.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.2 | 0.5 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.2 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.6 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.1 | 0.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.4 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 1.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 2.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 1.4 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 5.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 1.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.7 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 0.8 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 11.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.2 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 5.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.9 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.7 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 4.7 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.6 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.4 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.1 | 2.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.8 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.6 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 6.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.4 | GO:0042556 | cobinamide kinase activity(GO:0008819) phytol kinase activity(GO:0010276) phenol kinase activity(GO:0018720) cyclin-dependent protein kinase activating kinase regulator activity(GO:0019914) inositol tetrakisphosphate 2-kinase activity(GO:0032942) heptose 7-phosphate kinase activity(GO:0033785) aminoglycoside phosphotransferase activity(GO:0034071) eukaryotic elongation factor-2 kinase regulator activity(GO:0042556) eukaryotic elongation factor-2 kinase activator activity(GO:0042557) LPPG:FO 2-phospho-L-lactate transferase activity(GO:0043743) cytidine kinase activity(GO:0043771) glycerate 2-kinase activity(GO:0043798) (S)-lactate 2-kinase activity(GO:0043841) phosphoserine:homoserine phosphotransferase activity(GO:0043899) L-seryl-tRNA(Sec) kinase activity(GO:0043915) phosphocholine transferase activity(GO:0044605) GTP-dependent polynucleotide kinase activity(GO:0051735) farnesol kinase activity(GO:0052668) CTP:2-trans,-6-trans-farnesol kinase activity(GO:0052669) geraniol kinase activity(GO:0052670) geranylgeraniol kinase activity(GO:0052671) CTP:geranylgeraniol kinase activity(GO:0052672) prenol kinase activity(GO:0052673) 1-phosphatidylinositol-5-kinase activity(GO:0052810) 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) inositol 5-diphosphate pentakisphosphate 5-kinase activity(GO:0052836) inositol diphosphate tetrakisphosphate kinase activity(GO:0052839) |
| 0.0 | 1.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |