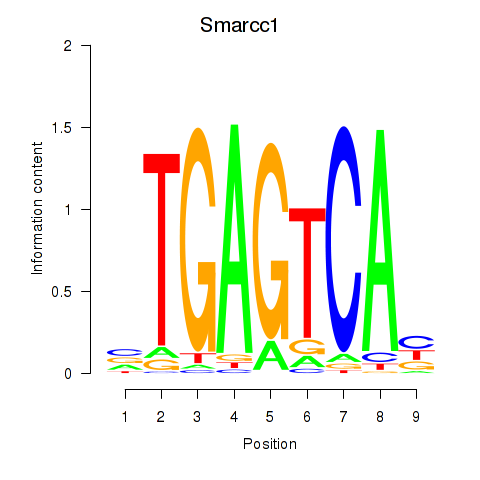
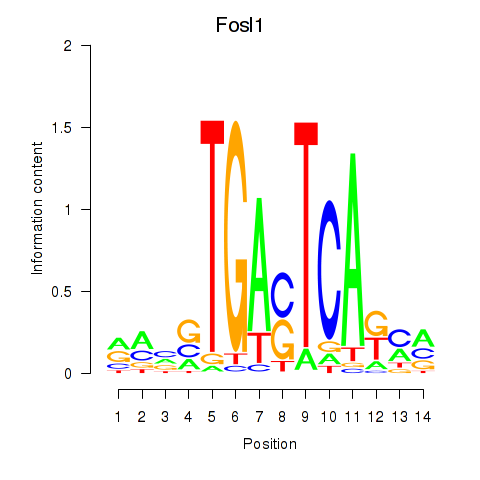
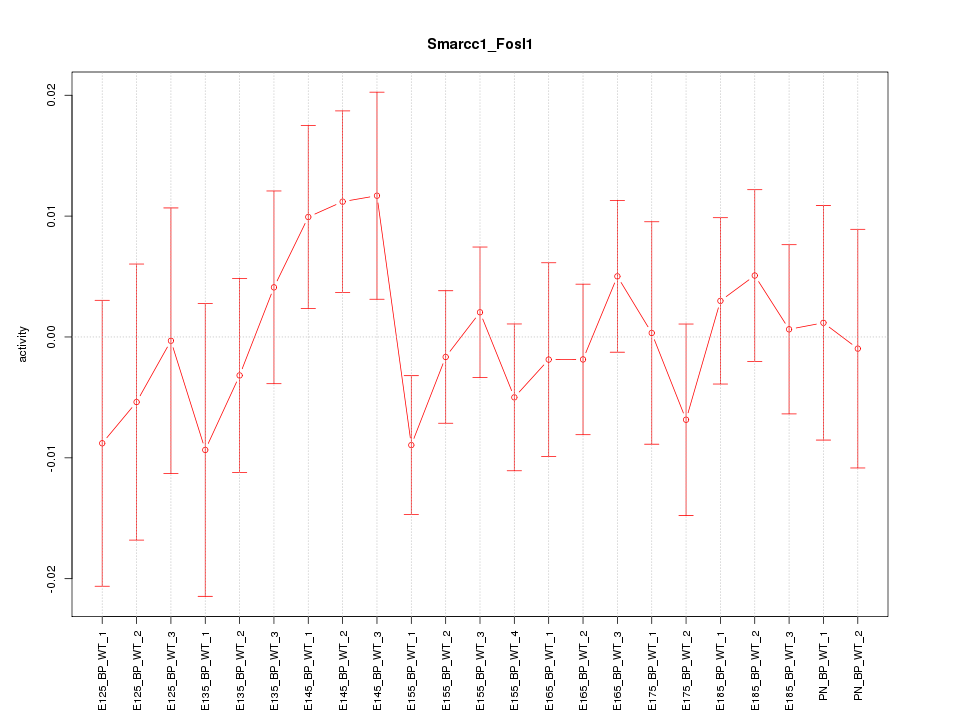
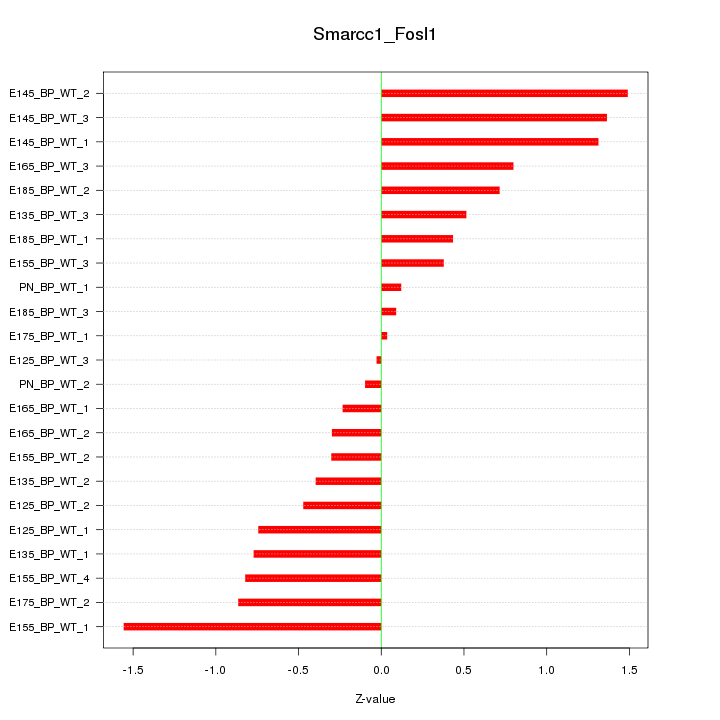
| 0.4 |
2.3 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.3 |
1.6 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.3 |
1.2 |
GO:1901204 |
regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.3 |
0.9 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.3 |
0.8 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.2 |
1.0 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 |
0.2 |
GO:0010751 |
negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.2 |
0.9 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 |
1.4 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.2 |
1.6 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 |
0.5 |
GO:0046370 |
fructose biosynthetic process(GO:0046370) |
| 0.1 |
0.6 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.1 |
0.7 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.6 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.4 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.1 |
0.4 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 |
0.8 |
GO:0046103 |
ADP biosynthetic process(GO:0006172) inosine biosynthetic process(GO:0046103) |
| 0.1 |
0.4 |
GO:0051031 |
tRNA transport(GO:0051031) |
| 0.1 |
0.8 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.1 |
0.8 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 |
0.4 |
GO:0043181 |
vacuolar sequestering(GO:0043181) |
| 0.1 |
0.1 |
GO:0043096 |
purine nucleobase salvage(GO:0043096) |
| 0.1 |
0.6 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.3 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 |
0.5 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.6 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.1 |
GO:0045763 |
negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.8 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.1 |
0.3 |
GO:0046032 |
ADP catabolic process(GO:0046032) |
| 0.1 |
0.3 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 |
0.3 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 |
0.7 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.3 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.2 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.1 |
0.5 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.6 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
0.1 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.1 |
0.4 |
GO:0008078 |
mesodermal cell migration(GO:0008078) |
| 0.1 |
0.4 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 |
0.6 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.3 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 |
0.9 |
GO:0051152 |
positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.1 |
0.7 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
0.3 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.1 |
0.3 |
GO:0031038 |
myosin II filament organization(GO:0031038) regulation of myosin II filament organization(GO:0043519) |
| 0.1 |
0.5 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 |
0.4 |
GO:1902031 |
regulation of NADP metabolic process(GO:1902031) |
| 0.1 |
0.2 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.1 |
0.3 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.2 |
GO:0009298 |
GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 |
0.1 |
GO:1902455 |
negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 |
0.3 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.1 |
0.1 |
GO:0035434 |
copper ion transmembrane transport(GO:0035434) |
| 0.1 |
0.2 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 |
0.8 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 |
0.2 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
0.4 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.9 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.3 |
GO:2000468 |
regulation of peroxidase activity(GO:2000468) |
| 0.1 |
0.6 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 |
0.3 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
0.2 |
GO:0019478 |
D-amino acid catabolic process(GO:0019478) |
| 0.1 |
0.2 |
GO:0039521 |
suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 |
0.2 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.3 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 |
0.3 |
GO:0042167 |
heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 |
0.1 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 0.1 |
0.4 |
GO:1903056 |
melanocyte migration(GO:0097324) regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 |
0.6 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.1 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
0.4 |
GO:0050965 |
detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 |
0.3 |
GO:1901252 |
regulation of intracellular transport of viral material(GO:1901252) |
| 0.1 |
0.2 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 |
0.4 |
GO:1903147 |
negative regulation of mitophagy(GO:1903147) |
| 0.1 |
0.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.3 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.2 |
GO:1904996 |
positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.1 |
0.3 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 |
0.1 |
GO:1902861 |
copper ion import into cell(GO:1902861) |
| 0.1 |
0.2 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.1 |
0.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 |
0.5 |
GO:0018916 |
nitrobenzene metabolic process(GO:0018916) |
| 0.1 |
0.3 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.1 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.1 |
0.3 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.4 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.2 |
GO:1904956 |
regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 |
0.2 |
GO:0002339 |
B cell selection(GO:0002339) |
| 0.1 |
0.5 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.3 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.2 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 |
0.2 |
GO:0070428 |
granuloma formation(GO:0002432) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 |
0.2 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.1 |
0.6 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 |
0.5 |
GO:0030049 |
muscle filament sliding(GO:0030049) |
| 0.1 |
0.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
0.3 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.1 |
0.1 |
GO:0019343 |
cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 0.0 |
0.2 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.0 |
0.3 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
0.1 |
GO:1900041 |
intestinal D-glucose absorption(GO:0001951) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.0 |
0.1 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.0 |
0.5 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 |
0.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 |
0.2 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.0 |
0.2 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.0 |
0.2 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.0 |
0.2 |
GO:0090086 |
negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 |
0.1 |
GO:0071492 |
cellular response to UV-A(GO:0071492) |
| 0.0 |
0.1 |
GO:0006558 |
L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.0 |
0.2 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 |
0.4 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.6 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.0 |
0.2 |
GO:0072656 |
regulation of anion channel activity(GO:0010359) maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 |
0.1 |
GO:0009644 |
response to high light intensity(GO:0009644) |
| 0.0 |
0.1 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 |
0.1 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
1.0 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.2 |
GO:0042701 |
progesterone secretion(GO:0042701) |
| 0.0 |
1.7 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.5 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
0.2 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.1 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 |
0.3 |
GO:0051418 |
interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 |
0.1 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
1.7 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.2 |
GO:0006689 |
ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) |
| 0.0 |
0.1 |
GO:0044034 |
negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.3 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.2 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.0 |
0.3 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.0 |
0.6 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 |
0.7 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.3 |
GO:0071816 |
protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 |
0.1 |
GO:0038145 |
macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 |
1.4 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.0 |
0.1 |
GO:0019405 |
alditol catabolic process(GO:0019405) |
| 0.0 |
0.2 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.5 |
GO:0032808 |
lacrimal gland development(GO:0032808) |
| 0.0 |
0.1 |
GO:2000334 |
response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 |
0.3 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
0.2 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.1 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 |
0.2 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.0 |
0.3 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.0 |
0.1 |
GO:0000454 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 |
0.1 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.4 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
0.3 |
GO:0010878 |
cholesterol storage(GO:0010878) |
| 0.0 |
0.1 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 |
1.0 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.1 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) |
| 0.0 |
0.1 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.0 |
0.2 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 |
0.1 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.0 |
0.8 |
GO:0033198 |
response to ATP(GO:0033198) |
| 0.0 |
0.2 |
GO:0048549 |
positive regulation of pinocytosis(GO:0048549) |
| 0.0 |
0.5 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.2 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.1 |
GO:0060160 |
negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 |
0.1 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.1 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
0.5 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.0 |
0.3 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 |
0.2 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.1 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.3 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.1 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.1 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 |
0.2 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.1 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 |
0.1 |
GO:0097680 |
double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 |
0.1 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.0 |
0.1 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.2 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.3 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.1 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.0 |
0.3 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.1 |
GO:0098912 |
smooth muscle contraction involved in micturition(GO:0060083) membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 |
0.1 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 |
0.1 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.0 |
0.1 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 |
0.3 |
GO:0046051 |
UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 |
0.2 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 |
0.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.1 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
0.2 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.3 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 |
0.2 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.1 |
GO:0070682 |
proteasome regulatory particle assembly(GO:0070682) |
| 0.0 |
0.3 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.1 |
GO:0070417 |
cellular response to cold(GO:0070417) |
| 0.0 |
0.1 |
GO:0019388 |
galactose catabolic process(GO:0019388) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.1 |
GO:0051490 |
negative regulation of filopodium assembly(GO:0051490) |
| 0.0 |
0.3 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.5 |
GO:0060707 |
trophoblast giant cell differentiation(GO:0060707) |
| 0.0 |
1.0 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.2 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.1 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 |
0.1 |
GO:0030223 |
neutrophil differentiation(GO:0030223) |
| 0.0 |
0.3 |
GO:0060561 |
apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 |
0.2 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.0 |
0.1 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.2 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.1 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 |
0.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
0.2 |
GO:0070627 |
ferrous iron import(GO:0070627) |
| 0.0 |
0.1 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.0 |
0.1 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.0 |
0.2 |
GO:0006750 |
glutathione biosynthetic process(GO:0006750) |
| 0.0 |
0.1 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.0 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 |
0.3 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.2 |
GO:0015816 |
glycine transport(GO:0015816) |
| 0.0 |
0.3 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.1 |
GO:0006538 |
glutamate catabolic process(GO:0006538) |
| 0.0 |
0.1 |
GO:0072110 |
glomerular mesangial cell proliferation(GO:0072110) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.2 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.1 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 |
0.0 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.0 |
0.8 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.0 |
0.2 |
GO:0044406 |
adhesion of symbiont to host(GO:0044406) |
| 0.0 |
0.1 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.0 |
0.2 |
GO:0086103 |
G-protein coupled receptor signaling pathway involved in heart process(GO:0086103) |
| 0.0 |
0.8 |
GO:0007193 |
adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 |
0.2 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 |
0.1 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.0 |
0.0 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.0 |
0.1 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 |
0.2 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.1 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 |
1.3 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.1 |
GO:0019673 |
GDP-mannose metabolic process(GO:0019673) |
| 0.0 |
0.1 |
GO:0045743 |
positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.1 |
GO:0045624 |
positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.0 |
0.2 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
0.1 |
GO:0008206 |
bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 |
0.1 |
GO:0006878 |
cellular copper ion homeostasis(GO:0006878) |
| 0.0 |
0.3 |
GO:0019835 |
cytolysis(GO:0019835) |
| 0.0 |
0.1 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 |
0.2 |
GO:0061615 |
glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.0 |
0.1 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.3 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.0 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.3 |
GO:0045540 |
regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 |
0.1 |
GO:2000416 |
regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.0 |
0.1 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.2 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 |
0.1 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 |
0.1 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.0 |
0.1 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.3 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.1 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.0 |
0.1 |
GO:0048681 |
negative regulation of axon regeneration(GO:0048681) |
| 0.0 |
0.1 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.0 |
0.1 |
GO:0032429 |
regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 |
0.1 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 |
0.1 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 |
0.1 |
GO:0031053 |
production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) |
| 0.0 |
0.1 |
GO:0033602 |
negative regulation of dopamine secretion(GO:0033602) |
| 0.0 |
0.2 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.0 |
0.7 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.1 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 |
0.1 |
GO:0009227 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) nucleotide-sugar catabolic process(GO:0009227) |
| 0.0 |
1.1 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.0 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.0 |
0.1 |
GO:0032364 |
oxygen homeostasis(GO:0032364) |
| 0.0 |
0.0 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 |
0.1 |
GO:0006776 |
vitamin A metabolic process(GO:0006776) |
| 0.0 |
0.0 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.1 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 |
0.0 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.0 |
0.2 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 |
0.3 |
GO:0001539 |
cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 |
0.2 |
GO:0071451 |
cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) |
| 0.0 |
0.2 |
GO:0039702 |
viral budding via host ESCRT complex(GO:0039702) |
| 0.0 |
0.1 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.1 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.0 |
0.0 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.0 |
0.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.0 |
0.1 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.2 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.0 |
0.0 |
GO:0042537 |
benzene-containing compound metabolic process(GO:0042537) |
| 0.0 |
0.0 |
GO:0051462 |
cortisol secretion(GO:0043400) regulation of cortisol secretion(GO:0051462) positive regulation of cortisol secretion(GO:0051464) |
| 0.0 |
0.1 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 |
0.1 |
GO:0006729 |
tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 |
0.1 |
GO:0051096 |
telomere assembly(GO:0032202) regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.2 |
GO:0072666 |
establishment of protein localization to vacuole(GO:0072666) |
| 0.0 |
0.2 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.0 |
GO:0070493 |
thrombin receptor signaling pathway(GO:0070493) |
| 0.0 |
0.1 |
GO:0002031 |
G-protein coupled receptor internalization(GO:0002031) |
| 0.0 |
0.1 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 |
0.1 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.1 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 |
0.2 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.2 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
0.4 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 |
0.1 |
GO:0018202 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) peptidyl-histidine modification(GO:0018202) |
| 0.0 |
0.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.1 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 |
0.1 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.2 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
0.3 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.0 |
GO:0035793 |
cell migration involved in kidney development(GO:0035787) cell migration involved in metanephros development(GO:0035788) metanephric mesenchymal cell migration(GO:0035789) positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of metanephric mesenchymal cell migration(GO:2000589) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 |
0.0 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.1 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.2 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0097034 |
mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 |
0.3 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.0 |
0.3 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.0 |
0.1 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.0 |
0.1 |
GO:0000291 |
nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.0 |
0.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 |
0.1 |
GO:0097286 |
iron ion import(GO:0097286) |
| 0.0 |
0.2 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.0 |
0.6 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
0.0 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.0 |
0.2 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.1 |
GO:0006266 |
DNA ligation(GO:0006266) |
| 0.0 |
0.1 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.0 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
0.1 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.2 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.0 |
0.1 |
GO:0036265 |
RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 |
0.1 |
GO:0030150 |
protein import into mitochondrial matrix(GO:0030150) |
| 0.0 |
0.1 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 |
0.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.3 |
GO:0010972 |
negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 |
0.0 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.1 |
GO:2000786 |
positive regulation of vacuole organization(GO:0044090) positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.2 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.0 |
GO:0043030 |
regulation of macrophage activation(GO:0043030) |
| 0.0 |
0.0 |
GO:0021759 |
globus pallidus development(GO:0021759) |