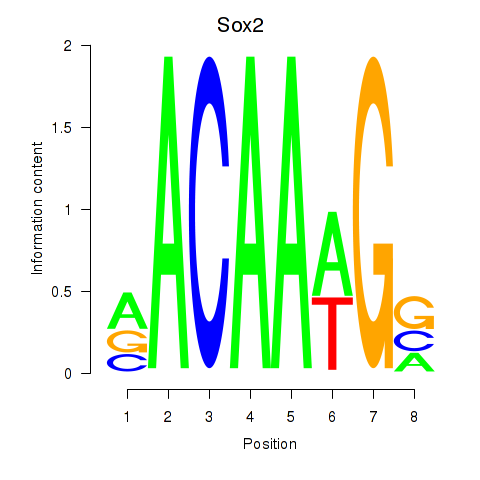
| 2.6 |
7.9 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.5 |
7.4 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 1.4 |
7.0 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 1.3 |
3.9 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 1.3 |
6.5 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.2 |
3.7 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 1.2 |
3.6 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.2 |
5.8 |
GO:0065001 |
specification of axis polarity(GO:0065001) |
| 1.2 |
4.6 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 1.1 |
1.1 |
GO:0009786 |
regulation of asymmetric cell division(GO:0009786) |
| 1.1 |
6.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.1 |
12.7 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 1.1 |
3.2 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.0 |
12.3 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.9 |
4.5 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.8 |
5.9 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.8 |
2.5 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.8 |
6.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.8 |
3.3 |
GO:0021986 |
epithalamus development(GO:0021538) habenula development(GO:0021986) general adaptation syndrome(GO:0051866) |
| 0.8 |
3.3 |
GO:2001015 |
negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.8 |
2.5 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.8 |
7.1 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.8 |
4.0 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.8 |
2.3 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) |
| 0.8 |
2.3 |
GO:1905065 |
hematopoietic stem cell migration(GO:0035701) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.7 |
4.4 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.7 |
2.2 |
GO:0072139 |
glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.7 |
2.0 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.7 |
2.7 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.7 |
3.3 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.6 |
2.6 |
GO:2000705 |
regulation of dense core granule biogenesis(GO:2000705) |
| 0.6 |
5.8 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.6 |
1.9 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.6 |
5.0 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.6 |
2.5 |
GO:0061626 |
pharyngeal arch artery morphogenesis(GO:0061626) |
| 0.6 |
10.4 |
GO:0010715 |
regulation of extracellular matrix disassembly(GO:0010715) |
| 0.6 |
1.8 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.6 |
0.6 |
GO:0045779 |
negative regulation of bone resorption(GO:0045779) |
| 0.6 |
1.7 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.6 |
2.3 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.6 |
1.7 |
GO:2000344 |
positive regulation of acrosome reaction(GO:2000344) |
| 0.5 |
2.2 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.5 |
8.3 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.5 |
5.7 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.5 |
3.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.5 |
2.0 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.5 |
1.4 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.5 |
1.4 |
GO:0046022 |
regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) axial mesoderm morphogenesis(GO:0048319) |
| 0.5 |
1.9 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.5 |
0.9 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.5 |
0.5 |
GO:0061324 |
canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.5 |
4.5 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.5 |
0.5 |
GO:0097491 |
trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.4 |
2.2 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 |
2.6 |
GO:1903753 |
negative regulation of p38MAPK cascade(GO:1903753) |
| 0.4 |
1.7 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.4 |
3.5 |
GO:0032625 |
interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.4 |
2.6 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 |
3.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.4 |
6.8 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 |
7.4 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.4 |
2.4 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.4 |
2.4 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.4 |
1.2 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.4 |
2.0 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.4 |
2.0 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.4 |
1.6 |
GO:0010838 |
positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.4 |
1.2 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.4 |
1.2 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.4 |
3.1 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.4 |
3.1 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.4 |
1.2 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.4 |
3.1 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.4 |
1.2 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.4 |
2.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.4 |
2.6 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.4 |
1.1 |
GO:0048211 |
Golgi vesicle docking(GO:0048211) |
| 0.4 |
3.3 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.4 |
0.7 |
GO:0048505 |
regulation of timing of cell differentiation(GO:0048505) |
| 0.4 |
3.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.4 |
2.9 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.4 |
1.4 |
GO:0031133 |
regulation of axon diameter(GO:0031133) |
| 0.4 |
1.1 |
GO:2000616 |
negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.3 |
1.0 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.3 |
1.0 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.3 |
1.4 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.3 |
1.3 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 |
1.0 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.3 |
3.5 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.3 |
0.9 |
GO:0070649 |
formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 |
2.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.3 |
1.2 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 |
0.9 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.3 |
1.2 |
GO:0098904 |
regulation of AV node cell action potential(GO:0098904) |
| 0.3 |
11.1 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.3 |
0.6 |
GO:1902995 |
regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.3 |
4.7 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.3 |
0.9 |
GO:1903334 |
positive regulation of protein folding(GO:1903334) |
| 0.3 |
0.3 |
GO:0042275 |
error-free postreplication DNA repair(GO:0042275) |
| 0.3 |
0.6 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.3 |
1.4 |
GO:0046684 |
response to insecticide(GO:0017085) response to pyrethroid(GO:0046684) |
| 0.3 |
4.6 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 |
1.1 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.3 |
0.8 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.3 |
0.6 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.3 |
1.7 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.3 |
1.4 |
GO:0061198 |
fungiform papilla formation(GO:0061198) |
| 0.3 |
1.1 |
GO:0030214 |
hyaluronan catabolic process(GO:0030214) |
| 0.3 |
0.3 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.3 |
6.1 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.3 |
1.0 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.3 |
0.8 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.3 |
1.0 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.3 |
2.3 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) |
| 0.2 |
1.5 |
GO:0010739 |
positive regulation of protein kinase A signaling(GO:0010739) |
| 0.2 |
1.5 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.2 |
4.9 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 |
1.2 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.2 |
1.7 |
GO:1904754 |
positive regulation of vascular associated smooth muscle cell migration(GO:1904754) |
| 0.2 |
4.3 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.2 |
0.9 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 |
0.7 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 |
0.7 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 |
0.7 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.2 |
1.3 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 |
6.2 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.2 |
2.9 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.2 |
2.9 |
GO:0048875 |
chemical homeostasis within a tissue(GO:0048875) |
| 0.2 |
0.9 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.2 |
0.9 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 |
1.1 |
GO:0046826 |
negative regulation of protein export from nucleus(GO:0046826) |
| 0.2 |
0.9 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 |
0.6 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.2 |
0.9 |
GO:1903966 |
monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 |
0.9 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.2 |
3.0 |
GO:0034377 |
plasma lipoprotein particle assembly(GO:0034377) |
| 0.2 |
0.4 |
GO:2000286 |
receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.2 |
2.1 |
GO:0035090 |
maintenance of apical/basal cell polarity(GO:0035090) |
| 0.2 |
0.2 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.2 |
0.4 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.2 |
3.5 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.2 |
3.3 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.2 |
0.6 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 |
1.6 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 |
2.2 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.2 |
0.6 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.2 |
0.6 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.2 |
0.8 |
GO:0072385 |
minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.2 |
0.8 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.2 |
0.8 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.2 |
0.8 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 |
5.7 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 |
1.5 |
GO:0072176 |
nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.2 |
0.9 |
GO:0046666 |
retinal cell programmed cell death(GO:0046666) |
| 0.2 |
0.7 |
GO:0010815 |
bradykinin catabolic process(GO:0010815) |
| 0.2 |
1.5 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
1.3 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.2 |
1.6 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.2 |
0.5 |
GO:1905154 |
negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.2 |
1.9 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.2 |
2.4 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.2 |
0.7 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.2 |
1.7 |
GO:0086036 |
regulation of cardiac muscle cell membrane potential(GO:0086036) |
| 0.2 |
0.5 |
GO:0002053 |
positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.2 |
13.0 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.2 |
0.3 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.2 |
1.0 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.2 |
0.3 |
GO:0071680 |
response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.2 |
3.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.2 |
0.2 |
GO:0060051 |
negative regulation of protein glycosylation(GO:0060051) |
| 0.2 |
0.5 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
1.8 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 |
0.9 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.2 |
1.7 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.2 |
2.0 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.2 |
0.8 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 |
2.7 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
1.9 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 |
0.4 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 |
0.4 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
1.4 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.4 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 |
0.9 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.5 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.1 |
0.7 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
0.4 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.3 |
GO:0015697 |
quaternary ammonium group transport(GO:0015697) |
| 0.1 |
0.1 |
GO:2000017 |
positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 |
0.4 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
2.7 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.1 |
0.9 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.1 |
GO:0007172 |
signal complex assembly(GO:0007172) |
| 0.1 |
2.4 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.1 |
0.4 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.1 |
1.4 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 0.1 |
1.6 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.1 |
2.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.1 |
1.0 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 |
0.8 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.1 |
0.5 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.1 |
2.9 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 |
0.6 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 |
1.1 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
1.1 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.1 |
0.7 |
GO:0033523 |
histone H2B ubiquitination(GO:0033523) |
| 0.1 |
0.8 |
GO:0046543 |
development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.1 |
0.1 |
GO:0060664 |
epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.1 |
0.1 |
GO:0060800 |
regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 |
2.5 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.1 |
1.1 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
2.6 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.1 |
3.8 |
GO:0043551 |
regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 |
2.0 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.1 |
0.8 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 |
2.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
4.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
1.2 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 |
0.9 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
0.3 |
GO:0036216 |
response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.1 |
1.2 |
GO:0002098 |
tRNA wobble uridine modification(GO:0002098) |
| 0.1 |
1.5 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
0.4 |
GO:1900028 |
negative regulation of ruffle assembly(GO:1900028) |
| 0.1 |
0.9 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.1 |
1.1 |
GO:0070914 |
UV-damage excision repair(GO:0070914) |
| 0.1 |
0.8 |
GO:0006265 |
DNA topological change(GO:0006265) |
| 0.1 |
0.3 |
GO:0003263 |
cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) regulation of secondary heart field cardioblast proliferation(GO:0003266) cell proliferation involved in heart morphogenesis(GO:0061323) positive regulation of secondary heart field cardioblast proliferation(GO:0072513) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) regulation of heart morphogenesis(GO:2000826) |
| 0.1 |
0.8 |
GO:0031639 |
plasminogen activation(GO:0031639) |
| 0.1 |
0.3 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
1.0 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.1 |
0.5 |
GO:0002553 |
histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) Golgi reassembly(GO:0090168) |
| 0.1 |
0.6 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
5.5 |
GO:0010508 |
positive regulation of autophagy(GO:0010508) |
| 0.1 |
0.8 |
GO:0033152 |
immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 |
0.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 |
0.1 |
GO:0002606 |
dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 |
1.3 |
GO:0060009 |
Sertoli cell development(GO:0060009) |
| 0.1 |
0.8 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
1.4 |
GO:0001967 |
suckling behavior(GO:0001967) |
| 0.1 |
1.4 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.7 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 |
1.1 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.1 |
1.9 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.1 |
0.7 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 |
0.2 |
GO:0006592 |
ornithine biosynthetic process(GO:0006592) |
| 0.1 |
0.5 |
GO:0018377 |
N-terminal protein myristoylation(GO:0006499) protein myristoylation(GO:0018377) |
| 0.1 |
1.1 |
GO:0035020 |
regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 |
0.4 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
0.6 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.2 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 |
1.4 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.1 |
1.6 |
GO:1902042 |
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.1 |
0.3 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.6 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 |
0.1 |
GO:0048745 |
smooth muscle tissue development(GO:0048745) |
| 0.1 |
0.2 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.1 |
0.4 |
GO:0072553 |
terminal button organization(GO:0072553) |
| 0.1 |
2.2 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.1 |
1.0 |
GO:0001570 |
vasculogenesis(GO:0001570) |
| 0.1 |
0.4 |
GO:0050912 |
detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.1 |
0.4 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.1 |
0.1 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.1 |
0.6 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.1 |
0.4 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.8 |
GO:0033275 |
actin-myosin filament sliding(GO:0033275) |
| 0.1 |
0.1 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.1 |
1.9 |
GO:0042552 |
myelination(GO:0042552) |
| 0.1 |
1.0 |
GO:0072600 |
establishment of protein localization to Golgi(GO:0072600) |
| 0.1 |
0.5 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.1 |
2.0 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.3 |
GO:0072369 |
regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.1 |
0.2 |
GO:0060487 |
lung epithelial cell differentiation(GO:0060487) |
| 0.1 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
1.2 |
GO:0045947 |
negative regulation of translational initiation(GO:0045947) |
| 0.1 |
0.6 |
GO:1903504 |
regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 |
1.1 |
GO:0060384 |
innervation(GO:0060384) |
| 0.1 |
2.0 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.1 |
0.2 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.1 |
0.6 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.1 |
1.1 |
GO:0007035 |
vacuolar acidification(GO:0007035) |
| 0.1 |
2.0 |
GO:0006378 |
mRNA polyadenylation(GO:0006378) |
| 0.1 |
1.6 |
GO:0008089 |
anterograde axonal transport(GO:0008089) |
| 0.1 |
1.2 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.1 |
0.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.1 |
0.2 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.1 |
0.6 |
GO:0021794 |
thalamus development(GO:0021794) |
| 0.1 |
1.0 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
1.9 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.1 |
1.4 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.1 |
0.6 |
GO:0019319 |
hexose biosynthetic process(GO:0019319) |
| 0.1 |
0.1 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.4 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.1 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 |
1.6 |
GO:0046488 |
phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 |
1.1 |
GO:0032526 |
response to retinoic acid(GO:0032526) |
| 0.1 |
0.7 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.3 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.7 |
GO:0090205 |
positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 |
1.0 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.1 |
0.4 |
GO:0044872 |
lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 |
0.3 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.7 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.7 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.8 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.2 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.2 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.5 |
GO:0006366 |
transcription from RNA polymerase II promoter(GO:0006366) |
| 0.0 |
0.6 |
GO:0032024 |
positive regulation of insulin secretion(GO:0032024) |
| 0.0 |
0.6 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.1 |
GO:0051785 |
positive regulation of mitotic nuclear division(GO:0045840) positive regulation of nuclear division(GO:0051785) |
| 0.0 |
1.2 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.4 |
GO:0001832 |
blastocyst growth(GO:0001832) |
| 0.0 |
1.0 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
0.9 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.0 |
0.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.2 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.1 |
GO:0000965 |
mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 |
2.1 |
GO:1901652 |
response to peptide(GO:1901652) |
| 0.0 |
0.1 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.1 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.0 |
0.6 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
1.6 |
GO:0031647 |
regulation of protein stability(GO:0031647) |
| 0.0 |
1.1 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
1.1 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.0 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 |
2.3 |
GO:0050868 |
negative regulation of T cell activation(GO:0050868) |
| 0.0 |
0.3 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.0 |
0.5 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 |
0.4 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
2.3 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
0.1 |
GO:0048239 |
negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 |
0.1 |
GO:0061428 |
embryonic heart tube left/right pattern formation(GO:0060971) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.2 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.7 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.1 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 |
0.6 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.3 |
GO:0035092 |
sperm chromatin condensation(GO:0035092) |
| 0.0 |
0.2 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.8 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.0 |
0.3 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.8 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
1.8 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.9 |
GO:0032147 |
activation of protein kinase activity(GO:0032147) |
| 0.0 |
0.6 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.1 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.0 |
0.3 |
GO:0001783 |
B cell apoptotic process(GO:0001783) |
| 0.0 |
0.1 |
GO:0033008 |
positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
2.1 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
0.2 |
GO:0036120 |
response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 |
0.2 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
2.1 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.2 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.2 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.0 |
0.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.2 |
GO:0060998 |
regulation of dendritic spine development(GO:0060998) |
| 0.0 |
0.3 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
2.2 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.4 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.0 |
0.2 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.5 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.1 |
GO:0045198 |
establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 |
0.2 |
GO:0090200 |
positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 |
0.2 |
GO:1903961 |
positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 |
0.4 |
GO:0007127 |
meiosis I(GO:0007127) |
| 0.0 |
0.1 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.0 |
0.2 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.4 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.0 |
0.0 |
GO:0002572 |
pro-T cell differentiation(GO:0002572) |
| 0.0 |
0.4 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.1 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.0 |
0.0 |
GO:0001708 |
cell fate specification(GO:0001708) |
| 0.0 |
2.7 |
GO:0048813 |
dendrite morphogenesis(GO:0048813) |
| 0.0 |
0.2 |
GO:0046337 |
phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 |
1.7 |
GO:0010506 |
regulation of autophagy(GO:0010506) |
| 0.0 |
3.3 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
1.0 |
GO:0018105 |
peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 |
0.1 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.5 |
GO:0070527 |
platelet aggregation(GO:0070527) |
| 0.0 |
0.7 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.1 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 |
0.2 |
GO:1903427 |
negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.0 |
0.9 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.3 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
1.5 |
GO:0010950 |
positive regulation of endopeptidase activity(GO:0010950) |
| 0.0 |
0.2 |
GO:0021535 |
cell migration in hindbrain(GO:0021535) |
| 0.0 |
0.1 |
GO:0006527 |
arginine catabolic process(GO:0006527) |
| 0.0 |
0.8 |
GO:0030148 |
sphingolipid biosynthetic process(GO:0030148) |
| 0.0 |
0.2 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.1 |
GO:0030206 |
chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 |
0.1 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 |
0.1 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.2 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.3 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.6 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.5 |
GO:0018279 |
peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.2 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.6 |
GO:0051290 |
protein heterotetramerization(GO:0051290) |
| 0.0 |
0.1 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.4 |
GO:0000082 |
G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 |
0.5 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.8 |
GO:0006457 |
protein folding(GO:0006457) |
| 0.0 |
0.7 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.0 |
0.4 |
GO:0097194 |
execution phase of apoptosis(GO:0097194) |
| 0.0 |
0.3 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.9 |
GO:0001843 |
neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 |
0.2 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.1 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.1 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |