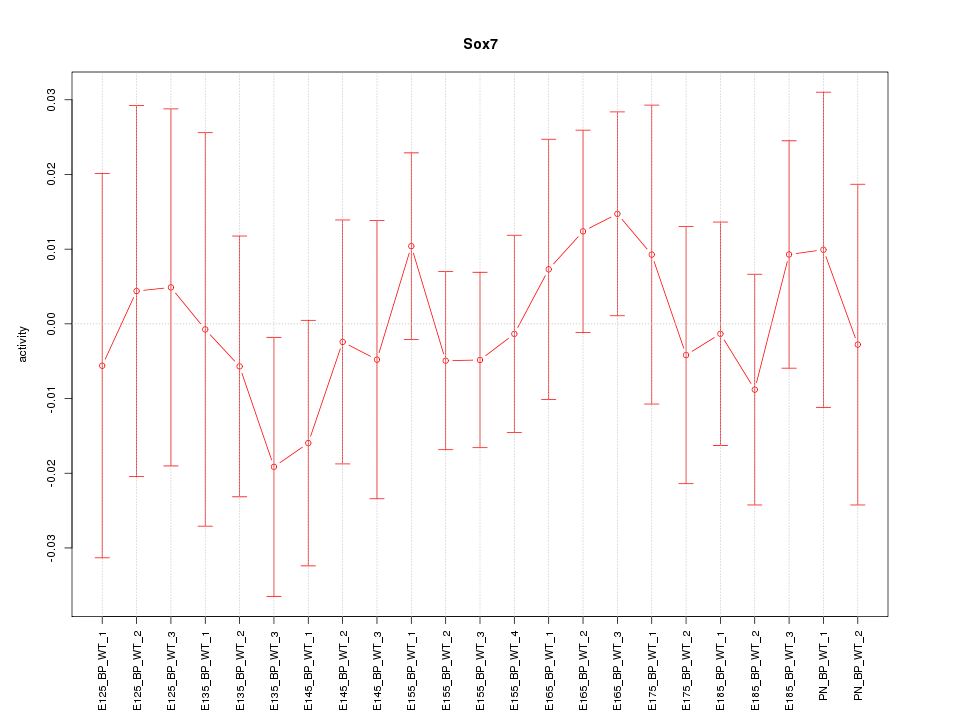
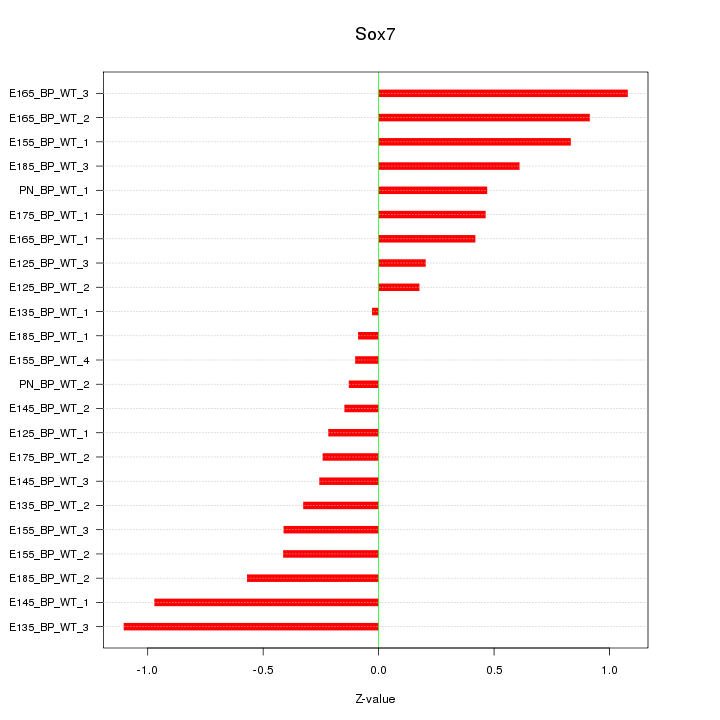
Motif ID: Sox7
Z-value: 0.548
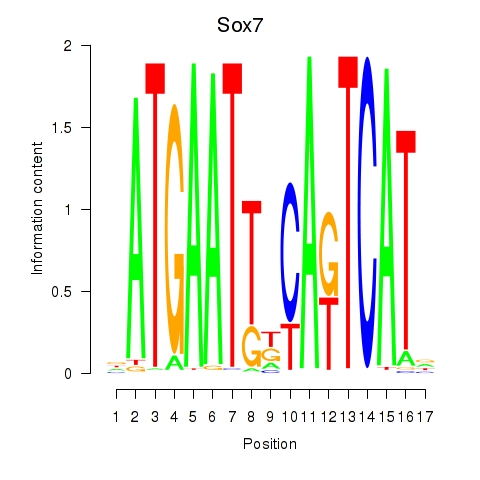
Transcription factors associated with Sox7:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sox7 | ENSMUSG00000063060.5 | Sox7 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox7 | mm10_v2_chr14_+_63943666_63943687 | 0.16 | 4.8e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001866 | NK T cell proliferation(GO:0001866) regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.1 | 0.2 | GO:0045472 | response to ether(GO:0045472) |
| 0.1 | 0.3 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.5 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 1.3 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.0 | 0.0 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 1.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |