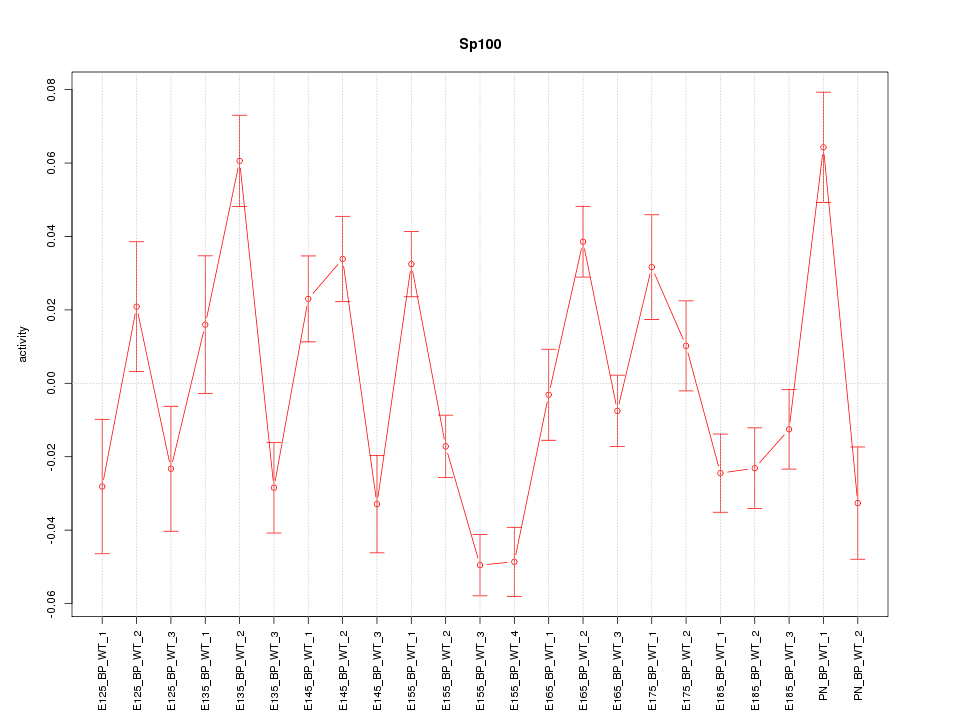
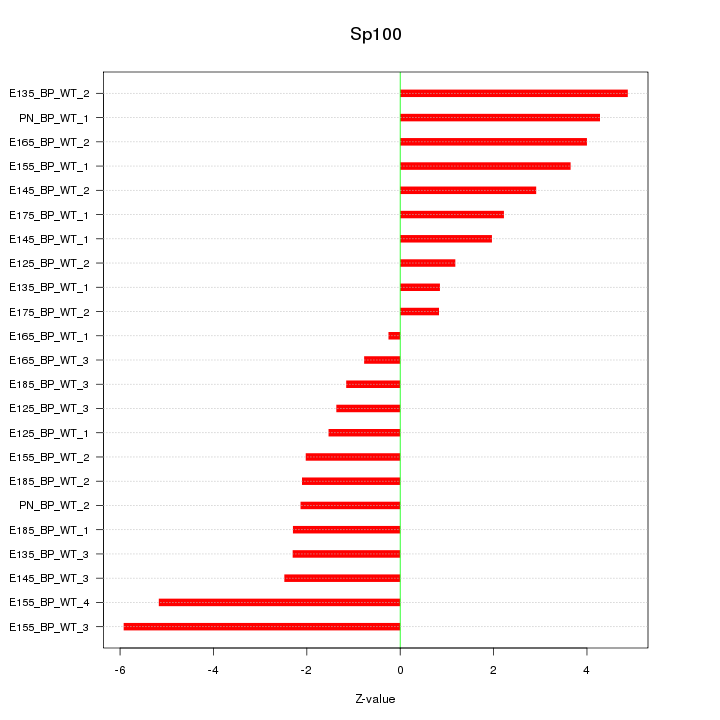
Motif ID: Sp100
Z-value: 2.869
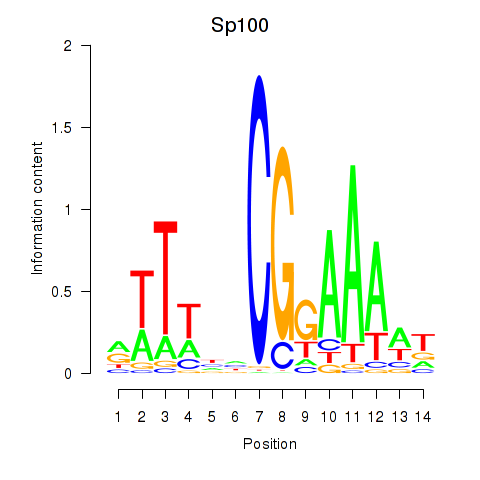
Transcription factors associated with Sp100:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Sp100 | ENSMUSG00000026222.10 | Sp100 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | mm10_v2_chr1_+_85650044_85650052 | 0.16 | 4.6e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 | 0.8 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.2 | 1.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.1 | 1.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.7 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.1 | 0.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.1 | 0.4 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.5 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 0.3 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 1.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.7 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.5 | GO:0010273 | detoxification of copper ion(GO:0010273) cellular response to zinc ion(GO:0071294) stress response to copper ion(GO:1990169) |
| 0.1 | 0.6 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.4 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.5 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 | 0.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.4 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:1903722 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.3 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.0 | 0.7 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2000828 | regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.0 | 0.2 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 | 0.3 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.0 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.3 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.0 | 0.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.4 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.4 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.5 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.1 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.5 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.7 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.7 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.2 | 0.6 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.5 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.1 | 0.6 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.1 | 0.3 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.6 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.0 | 0.5 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.6 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.7 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.0 | 1.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |