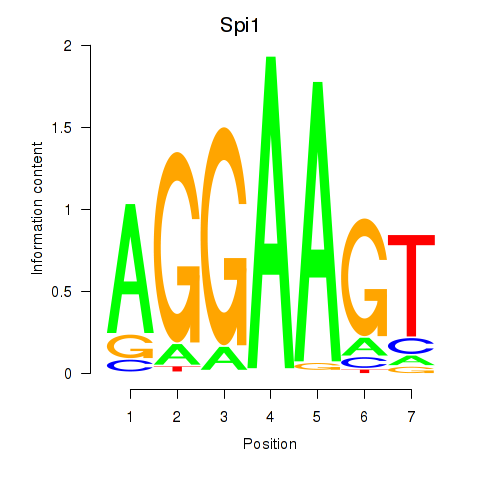
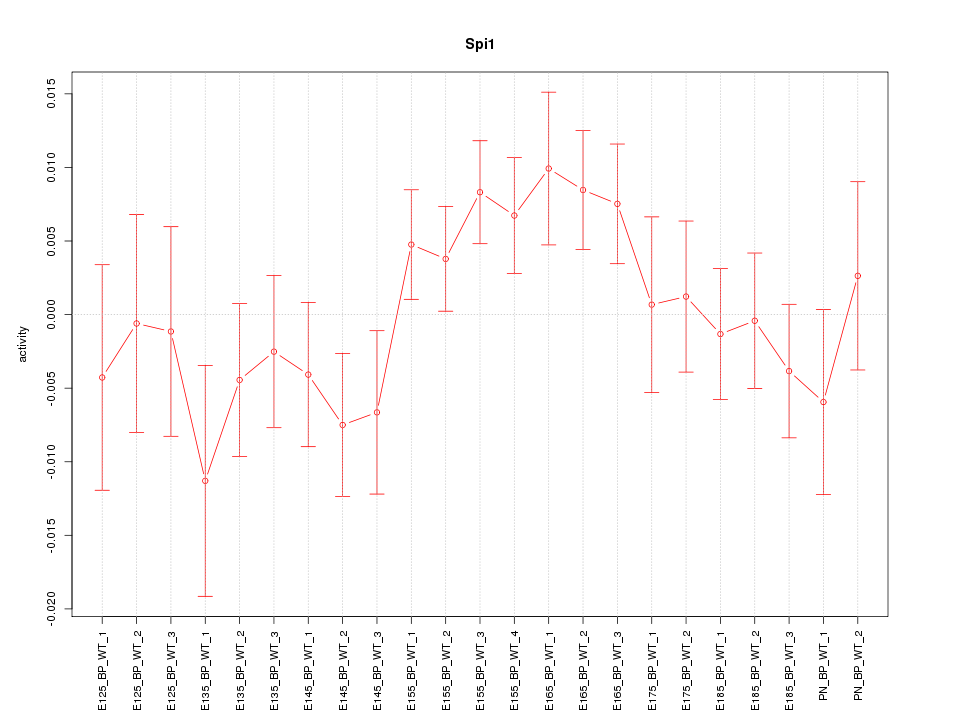
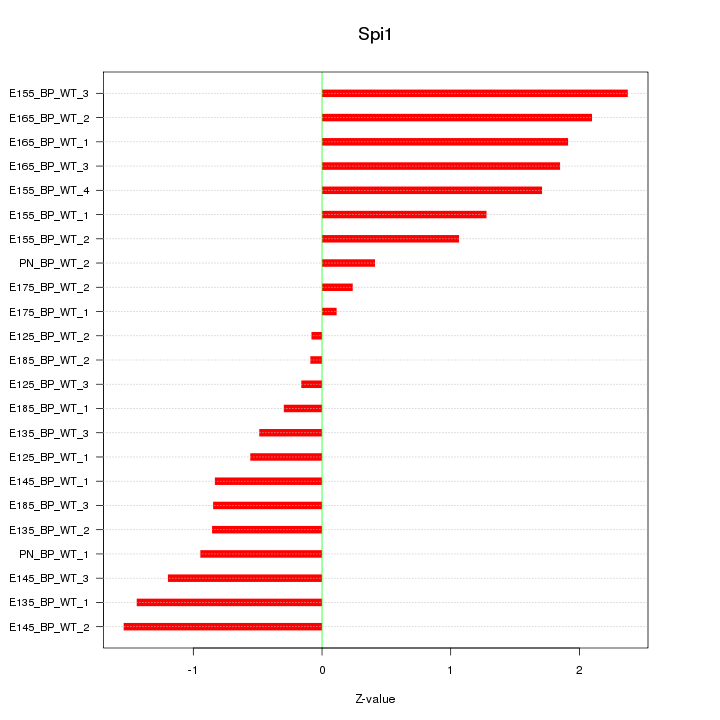
| 0.7 |
2.1 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.7 |
2.1 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.6 |
1.9 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.5 |
3.1 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.5 |
2.0 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.5 |
2.8 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 0.5 |
1.4 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.4 |
2.2 |
GO:0003213 |
cardiac right atrium morphogenesis(GO:0003213) |
| 0.4 |
1.2 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.4 |
1.6 |
GO:0006600 |
creatine metabolic process(GO:0006600) |
| 0.4 |
4.3 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.4 |
1.6 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.4 |
1.8 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.3 |
1.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 |
1.6 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.3 |
0.9 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.3 |
0.3 |
GO:0030201 |
heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.3 |
0.9 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.3 |
0.9 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.3 |
1.5 |
GO:0060690 |
epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.3 |
1.5 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.3 |
0.3 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.3 |
1.4 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 0.3 |
0.8 |
GO:0089700 |
protein kinase D signaling(GO:0089700) |
| 0.3 |
0.3 |
GO:1903279 |
regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 |
0.8 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.3 |
0.8 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.3 |
1.6 |
GO:0071883 |
activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.3 |
1.3 |
GO:0032962 |
positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.3 |
2.9 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.3 |
0.5 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.3 |
1.8 |
GO:1902669 |
positive regulation of axon extension involved in axon guidance(GO:0048842) positive regulation of axon guidance(GO:1902669) |
| 0.3 |
0.8 |
GO:1903054 |
negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 |
0.5 |
GO:0050713 |
negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.2 |
1.2 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.2 |
0.5 |
GO:1902566 |
regulation of eosinophil activation(GO:1902566) |
| 0.2 |
0.2 |
GO:0023021 |
termination of signal transduction(GO:0023021) |
| 0.2 |
0.9 |
GO:0021775 |
smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.2 |
0.7 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.2 |
0.7 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 |
1.1 |
GO:1902894 |
negative regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902894) |
| 0.2 |
1.8 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 |
0.2 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 |
2.8 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.2 |
0.9 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
0.2 |
GO:0003284 |
septum primum development(GO:0003284) |
| 0.2 |
1.5 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
0.6 |
GO:0010636 |
positive regulation of mitochondrial fusion(GO:0010636) |
| 0.2 |
4.6 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.2 |
0.6 |
GO:1905154 |
negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.2 |
0.6 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.2 |
0.6 |
GO:0021852 |
pyramidal neuron migration(GO:0021852) |
| 0.2 |
0.4 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.2 |
1.0 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 |
0.6 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.2 |
0.6 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.2 |
0.9 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 |
0.2 |
GO:0003415 |
chondrocyte hypertrophy(GO:0003415) |
| 0.2 |
1.3 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.2 |
0.4 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.2 |
1.8 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 |
0.5 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 |
1.5 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.2 |
1.2 |
GO:0032494 |
response to peptidoglycan(GO:0032494) |
| 0.2 |
0.3 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 |
0.5 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.2 |
0.6 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.2 |
0.5 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.2 |
1.3 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.2 |
0.5 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.2 |
0.6 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.2 |
0.8 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.2 |
0.5 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 |
0.5 |
GO:0036118 |
hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 |
1.5 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.1 |
0.3 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 |
0.4 |
GO:2000819 |
regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 |
0.1 |
GO:0090298 |
negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.1 |
0.4 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.1 |
0.6 |
GO:0021847 |
ventricular zone neuroblast division(GO:0021847) |
| 0.1 |
0.4 |
GO:2000851 |
positive regulation of cortisol secretion(GO:0051464) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.1 |
0.7 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.1 |
GO:2000338 |
toll-like receptor 5 signaling pathway(GO:0034146) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 |
0.7 |
GO:0044314 |
protein K27-linked ubiquitination(GO:0044314) |
| 0.1 |
1.4 |
GO:0070574 |
cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 |
0.3 |
GO:1903371 |
regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 |
2.2 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.1 |
0.6 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 |
0.4 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
1.1 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 |
0.7 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.1 |
0.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
0.9 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.1 |
0.4 |
GO:0060399 |
positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 |
1.2 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.1 |
0.4 |
GO:0036228 |
protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 |
0.4 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.4 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 |
0.5 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.1 |
0.3 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
1.9 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.8 |
GO:2001275 |
positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 |
0.4 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.1 |
0.5 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.3 |
GO:1903232 |
melanosome assembly(GO:1903232) |
| 0.1 |
0.4 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 |
0.4 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
1.1 |
GO:0060872 |
semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.1 |
0.9 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
1.0 |
GO:0048625 |
myoblast fate commitment(GO:0048625) |
| 0.1 |
0.4 |
GO:1904211 |
membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 |
0.8 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.1 |
GO:0042359 |
vitamin D metabolic process(GO:0042359) |
| 0.1 |
0.6 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.1 |
0.4 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.1 |
0.9 |
GO:0002318 |
myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 |
0.3 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.1 |
0.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
1.7 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
0.3 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.1 |
0.2 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 |
1.0 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.6 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
0.6 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) regulation of axon diameter(GO:0031133) interneuron migration(GO:1904936) |
| 0.1 |
0.6 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 |
0.2 |
GO:0061738 |
late endosomal microautophagy(GO:0061738) |
| 0.1 |
0.4 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.7 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.2 |
GO:0061511 |
centriole elongation(GO:0061511) |
| 0.1 |
0.8 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 |
0.9 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.5 |
GO:0033227 |
dsRNA transport(GO:0033227) |
| 0.1 |
0.7 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.3 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.1 |
0.4 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.1 |
0.4 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
1.3 |
GO:1902285 |
semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) |
| 0.1 |
1.1 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.1 |
1.3 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 |
0.7 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.1 |
1.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.7 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.9 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.3 |
GO:2000416 |
regulation of eosinophil migration(GO:2000416) positive regulation of eosinophil migration(GO:2000418) |
| 0.1 |
3.3 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.4 |
GO:1990743 |
protein sialylation(GO:1990743) |
| 0.1 |
0.2 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 |
1.2 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.8 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 |
1.1 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 |
0.3 |
GO:0033278 |
cell proliferation in midbrain(GO:0033278) |
| 0.1 |
0.3 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.1 |
0.4 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.1 |
1.4 |
GO:0097062 |
dendritic spine maintenance(GO:0097062) |
| 0.1 |
0.4 |
GO:1900368 |
regulation of RNA interference(GO:1900368) |
| 0.1 |
0.3 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.4 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
0.3 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.1 |
0.8 |
GO:0071763 |
nuclear membrane organization(GO:0071763) |
| 0.1 |
0.8 |
GO:0060059 |
embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 |
0.5 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.3 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.1 |
0.4 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 |
0.3 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.1 |
0.3 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 |
0.9 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.1 |
0.2 |
GO:0002756 |
MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 |
0.5 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
0.3 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 |
0.5 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.4 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.1 |
0.2 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.1 |
0.1 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.6 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.2 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 |
0.3 |
GO:0033030 |
negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 |
0.2 |
GO:2000256 |
positive regulation of male germ cell proliferation(GO:2000256) |
| 0.1 |
0.6 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.1 |
0.2 |
GO:0090289 |
regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
0.2 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
0.2 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.1 |
0.3 |
GO:1900157 |
regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.1 |
0.1 |
GO:0000972 |
transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.1 |
0.2 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.4 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.5 |
GO:0061668 |
mitochondrial ribosome assembly(GO:0061668) |
| 0.1 |
0.3 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.1 |
0.6 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 |
0.3 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.5 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.1 |
1.0 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.1 |
0.6 |
GO:0080009 |
mRNA methylation(GO:0080009) |
| 0.1 |
0.8 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
2.9 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
0.2 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.8 |
GO:1902170 |
cellular response to reactive nitrogen species(GO:1902170) |
| 0.1 |
0.2 |
GO:1902965 |
protein localization to early endosome(GO:1902946) regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 |
0.2 |
GO:1990046 |
positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) positive regulation of mitochondrial DNA metabolic process(GO:1901860) stress-induced mitochondrial fusion(GO:1990046) |
| 0.1 |
0.2 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.1 |
0.2 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 |
0.3 |
GO:0046477 |
glycosylceramide catabolic process(GO:0046477) |
| 0.1 |
0.7 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.1 |
0.1 |
GO:0060971 |
embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 |
0.2 |
GO:1903903 |
regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 |
1.0 |
GO:0061298 |
retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 |
0.2 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.1 |
GO:0097105 |
presynaptic membrane assembly(GO:0097105) |
| 0.1 |
1.3 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
0.1 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 |
1.3 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.1 |
0.6 |
GO:0032516 |
positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 |
0.3 |
GO:0090024 |
negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 |
0.3 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.1 |
0.3 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.2 |
GO:0021691 |
cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.1 |
0.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.3 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.1 |
0.3 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.1 |
0.6 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.1 |
0.3 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 |
0.5 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.1 |
0.4 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.1 |
0.2 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
0.4 |
GO:0035405 |
histone-threonine phosphorylation(GO:0035405) |
| 0.1 |
0.2 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.1 |
0.1 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 |
0.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.1 |
0.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.1 |
0.1 |
GO:0034141 |
positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.1 |
0.3 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.1 |
0.5 |
GO:0046459 |
short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 |
1.2 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.5 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.1 |
0.6 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.1 |
0.8 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.3 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.1 |
0.2 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.2 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.1 |
0.2 |
GO:0072673 |
lamellipodium morphogenesis(GO:0072673) |
| 0.1 |
0.2 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.3 |
GO:0070535 |
histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 |
0.2 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.1 |
0.4 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.4 |
GO:0030432 |
peristalsis(GO:0030432) |
| 0.1 |
0.2 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 |
0.3 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.1 |
0.8 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
0.3 |
GO:0006477 |
protein sulfation(GO:0006477) |
| 0.1 |
0.2 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.1 |
0.5 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
0.8 |
GO:0070816 |
phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.1 |
0.2 |
GO:0046697 |
decidualization(GO:0046697) |
| 0.1 |
0.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.2 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
0.5 |
GO:0061099 |
negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 |
0.8 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 |
0.6 |
GO:0003413 |
chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.1 |
0.4 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
1.0 |
GO:0045725 |
positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.1 |
0.6 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) regulation of membrane depolarization during action potential(GO:0098902) regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900825) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.3 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.1 |
0.7 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.1 |
0.3 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 |
0.2 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.1 |
0.4 |
GO:0052646 |
alditol phosphate metabolic process(GO:0052646) |
| 0.1 |
0.3 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
0.1 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 |
0.3 |
GO:0070447 |
positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) |
| 0.1 |
0.4 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.1 |
GO:1904398 |
regulation of synaptic growth at neuromuscular junction(GO:0008582) regulation of sodium ion export(GO:1903273) regulation of sodium ion export from cell(GO:1903276) regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 |
0.3 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.3 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.1 |
GO:0042790 |
transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 |
0.1 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 |
0.0 |
GO:2000812 |
regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 |
0.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
0.2 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.4 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 |
0.1 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.2 |
GO:0090330 |
regulation of platelet aggregation(GO:0090330) |
| 0.0 |
0.2 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 |
0.1 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 |
0.0 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.0 |
0.2 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.3 |
GO:0097011 |
cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 |
0.4 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.0 |
0.7 |
GO:0003334 |
keratinocyte development(GO:0003334) |
| 0.0 |
0.2 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.2 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
0.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 |
0.0 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.0 |
0.4 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0006154 |
adenosine catabolic process(GO:0006154) |
| 0.0 |
0.9 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.1 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.0 |
0.5 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 |
0.2 |
GO:1900169 |
regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 |
0.1 |
GO:0032964 |
collagen biosynthetic process(GO:0032964) |
| 0.0 |
0.5 |
GO:0090051 |
negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
| 0.0 |
0.2 |
GO:0071894 |
histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.0 |
0.4 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 |
0.2 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.0 |
0.2 |
GO:0090091 |
positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 |
0.9 |
GO:0022401 |
desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.0 |
0.0 |
GO:1904251 |
regulation of bile acid metabolic process(GO:1904251) |
| 0.0 |
0.1 |
GO:1902202 |
regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 |
0.1 |
GO:0038203 |
TORC2 signaling(GO:0038203) |
| 0.0 |
0.8 |
GO:0006896 |
Golgi to vacuole transport(GO:0006896) |
| 0.0 |
0.2 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.2 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 |
0.5 |
GO:0060831 |
smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 |
0.7 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 |
0.2 |
GO:0016557 |
peroxisome membrane biogenesis(GO:0016557) |
| 0.0 |
0.1 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 |
0.2 |
GO:0007039 |
protein catabolic process in the vacuole(GO:0007039) |
| 0.0 |
0.1 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 |
0.1 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 |
0.3 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.1 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.0 |
0.3 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.4 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.0 |
0.4 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 |
0.0 |
GO:2000484 |
positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.0 |
0.4 |
GO:0060353 |
cell adhesion molecule production(GO:0060352) regulation of cell adhesion molecule production(GO:0060353) |
| 0.0 |
0.0 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 |
0.2 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.1 |
GO:0046122 |
purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 |
0.2 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 |
0.1 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.0 |
0.8 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.2 |
GO:0060330 |
regulation of response to interferon-gamma(GO:0060330) |
| 0.0 |
0.1 |
GO:0006026 |
aminoglycan catabolic process(GO:0006026) |
| 0.0 |
0.2 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.0 |
0.1 |
GO:0002158 |
osteoclast proliferation(GO:0002158) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.0 |
GO:0010873 |
positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 |
0.2 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.0 |
0.2 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.0 |
0.6 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
1.0 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.0 |
0.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 |
1.1 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.0 |
0.2 |
GO:0031114 |
regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.2 |
GO:0070861 |
regulation of protein exit from endoplasmic reticulum(GO:0070861) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.2 |
GO:0034720 |
histone H3-K4 demethylation(GO:0034720) |
| 0.0 |
0.1 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.0 |
0.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 |
0.3 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.1 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.0 |
0.1 |
GO:0060437 |
lung growth(GO:0060437) |
| 0.0 |
0.1 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 |
0.1 |
GO:0070124 |
mitochondrial translational initiation(GO:0070124) |
| 0.0 |
0.1 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.0 |
0.2 |
GO:2001023 |
regulation of response to drug(GO:2001023) |
| 0.0 |
0.1 |
GO:0031086 |
nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 |
0.4 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 |
0.0 |
GO:1903984 |
positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 |
0.2 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 |
0.1 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.2 |
GO:0032196 |
transposition(GO:0032196) |
| 0.0 |
0.2 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.1 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.1 |
GO:0000350 |
generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 |
0.2 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.3 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.0 |
0.2 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.1 |
GO:2000402 |
negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.3 |
GO:2000505 |
regulation of energy homeostasis(GO:2000505) |
| 0.0 |
0.2 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.0 |
0.6 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.2 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
0.4 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
1.4 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.2 |
GO:0006283 |
transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:2000650 |
negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 |
0.2 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) positive regulation of superoxide anion generation(GO:0032930) |
| 0.0 |
0.1 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 |
0.9 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.1 |
GO:0090245 |
axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 |
0.8 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:0060294 |
cilium movement involved in cell motility(GO:0060294) |
| 0.0 |
0.6 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.1 |
GO:0055118 |
negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.0 |
0.3 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.1 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.5 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 |
0.4 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.2 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 |
0.1 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.0 |
0.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 |
0.3 |
GO:0061157 |
mRNA destabilization(GO:0061157) |
| 0.0 |
0.1 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.0 |
0.1 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 |
0.1 |
GO:0000389 |
mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 |
0.5 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.4 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.0 |
0.1 |
GO:0090283 |
regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 |
0.1 |
GO:0010728 |
regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 |
0.1 |
GO:0048241 |
epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.0 |
1.3 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
0.2 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.2 |
GO:1900078 |
positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 |
0.2 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.2 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.0 |
0.2 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.1 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 |
0.1 |
GO:0035493 |
SNARE complex assembly(GO:0035493) |
| 0.0 |
0.1 |
GO:0006384 |
transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 |
0.2 |
GO:0002726 |
positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 |
0.0 |
GO:0032472 |
Golgi calcium ion transport(GO:0032472) |
| 0.0 |
0.1 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.1 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 |
0.1 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.1 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 |
0.2 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
0.2 |
GO:0001773 |
myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.5 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.0 |
GO:0019805 |
quinolinate biosynthetic process(GO:0019805) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.1 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.1 |
GO:0006360 |
transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 |
0.1 |
GO:2000352 |
negative regulation of endothelial cell apoptotic process(GO:2000352) |
| 0.0 |
0.3 |
GO:0061302 |
smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 |
0.2 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.0 |
GO:0031914 |
negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 |
0.0 |
GO:2000370 |
positive regulation of clathrin-mediated endocytosis(GO:2000370) |
| 0.0 |
0.1 |
GO:0009212 |
dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.0 |
0.2 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.0 |
0.1 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.0 |
0.0 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.4 |
GO:1901533 |
negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 |
0.3 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.0 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 |
0.1 |
GO:0032329 |
serine transport(GO:0032329) |
| 0.0 |
0.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.0 |
0.0 |
GO:0034134 |
toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 |
0.1 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.0 |
0.6 |
GO:0016601 |
Rac protein signal transduction(GO:0016601) |
| 0.0 |
0.3 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) |
| 0.0 |
0.0 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.0 |
0.6 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.1 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 |
0.3 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.3 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.1 |
GO:1902065 |
response to L-glutamate(GO:1902065) |
| 0.0 |
0.4 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.0 |
GO:0031282 |
regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 |
0.3 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.0 |
0.1 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.3 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
1.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.2 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 |
0.1 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 |
0.2 |
GO:0090503 |
RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 |
0.1 |
GO:0036462 |
TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.1 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.0 |
0.0 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.0 |
0.0 |
GO:0097278 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0070070 |
proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 |
0.1 |
GO:0033233 |
regulation of protein sumoylation(GO:0033233) |
| 0.0 |
0.7 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.0 |
0.1 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.2 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 |
0.2 |
GO:0009396 |
folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 |
0.3 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.0 |
0.1 |
GO:0001991 |
regulation of systemic arterial blood pressure by circulatory renin-angiotensin(GO:0001991) |
| 0.0 |
0.1 |
GO:0071800 |
podosome assembly(GO:0071800) |
| 0.0 |
0.9 |
GO:0007632 |
visual behavior(GO:0007632) |
| 0.0 |
0.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.1 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
0.1 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.0 |
0.3 |
GO:0001921 |
positive regulation of receptor recycling(GO:0001921) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.0 |
GO:0060686 |
regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 |
0.1 |
GO:0006273 |
lagging strand elongation(GO:0006273) |
| 0.0 |
0.1 |
GO:0090286 |
cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 |
0.1 |
GO:0072348 |
sulfur compound transport(GO:0072348) |
| 0.0 |
0.2 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.0 |
GO:0008355 |
olfactory learning(GO:0008355) |
| 0.0 |
0.3 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.0 |
GO:0045656 |
negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 |
0.0 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 |
0.1 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.2 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.0 |
0.1 |
GO:0097341 |
inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 |
0.2 |
GO:0006509 |
membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 |
0.2 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.0 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.0 |
0.2 |
GO:0034629 |
cellular protein complex localization(GO:0034629) |
| 0.0 |
0.1 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.1 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 |
0.1 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.1 |
GO:0060136 |
embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 |
0.6 |
GO:0030641 |
regulation of cellular pH(GO:0030641) |
| 0.0 |
0.3 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.2 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.1 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.3 |
GO:0040019 |
positive regulation of embryonic development(GO:0040019) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.1 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 |
0.1 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.5 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:0000289 |
nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 |
0.0 |
GO:0021508 |
floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 |
0.2 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.0 |
0.1 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.3 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
0.1 |
GO:0070126 |
mitochondrial translational termination(GO:0070126) |
| 0.0 |
0.4 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:0016180 |
snRNA processing(GO:0016180) |
| 0.0 |
0.0 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0007000 |
nucleolus organization(GO:0007000) |
| 0.0 |
0.1 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 |
0.1 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 |
0.1 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.0 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 |
0.9 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
0.1 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.0 |
0.1 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.0 |
0.4 |
GO:0043124 |
negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 |
0.3 |
GO:0006829 |
zinc II ion transport(GO:0006829) |
| 0.0 |
0.0 |
GO:1903391 |
regulation of adherens junction organization(GO:1903391) |
| 0.0 |
0.1 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.2 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 |
0.1 |
GO:0031584 |
activation of phospholipase D activity(GO:0031584) |
| 0.0 |
0.0 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 |
0.1 |
GO:0040036 |
regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 |
0.0 |
GO:0070162 |
adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 |
0.0 |
GO:0051561 |
positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 |
0.1 |
GO:0072337 |
modified amino acid transport(GO:0072337) |
| 0.0 |
0.1 |
GO:0031295 |
lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 |
0.0 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.4 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.1 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.0 |
0.1 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.3 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.0 |
0.0 |
GO:0070344 |
fat cell proliferation(GO:0070341) regulation of fat cell proliferation(GO:0070344) negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 |
0.1 |
GO:0060148 |
positive regulation of posttranscriptional gene silencing(GO:0060148) |
| 0.0 |
0.8 |
GO:0007030 |
Golgi organization(GO:0007030) |
| 0.0 |
0.2 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.0 |
0.0 |
GO:0006435 |
threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 |
0.1 |
GO:0034446 |
substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 |
0.1 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.1 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |