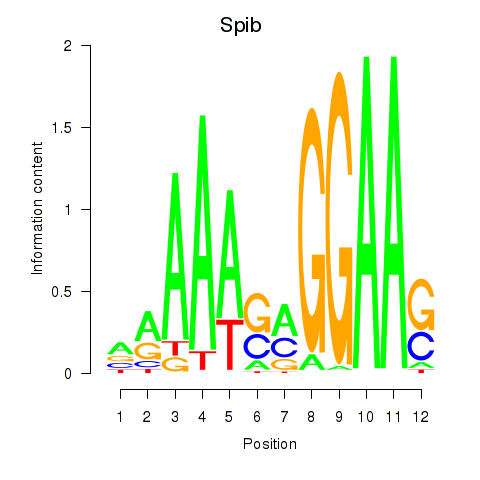
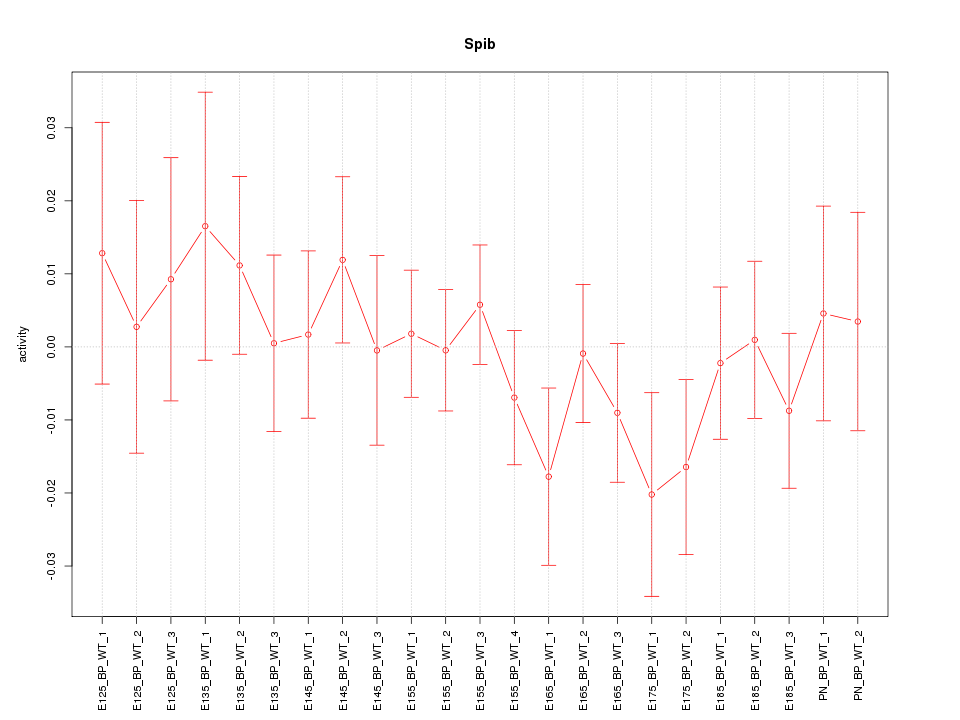
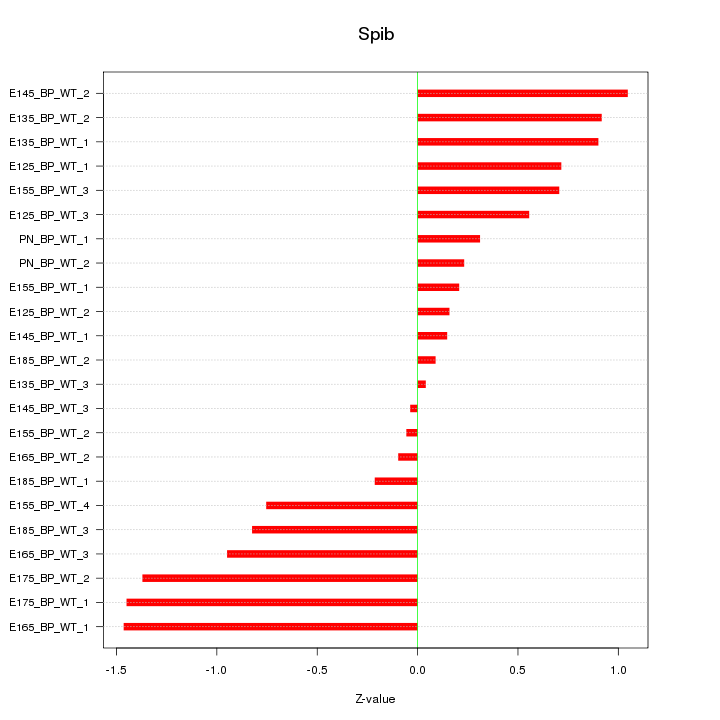
| 1.0 |
3.9 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.8 |
5.1 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.8 |
2.3 |
GO:0071649 |
regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) positive regulation of somatostatin secretion(GO:0090274) |
| 0.5 |
1.6 |
GO:0002538 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) |
| 0.5 |
2.5 |
GO:1901164 |
negative regulation of trophoblast cell migration(GO:1901164) |
| 0.5 |
2.4 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.4 |
2.3 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.3 |
1.4 |
GO:1903984 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.3 |
1.0 |
GO:1904059 |
regulation of locomotor rhythm(GO:1904059) |
| 0.3 |
0.9 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.2 |
0.7 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.2 |
0.7 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 |
0.6 |
GO:0070315 |
G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.2 |
0.7 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.2 |
0.5 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
0.4 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.4 |
GO:0033087 |
negative regulation of immature T cell proliferation(GO:0033087) |
| 0.1 |
0.4 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.1 |
1.8 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.6 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.8 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.6 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 |
0.3 |
GO:0048818 |
embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 0.1 |
0.3 |
GO:0001928 |
regulation of exocyst assembly(GO:0001928) |
| 0.1 |
1.1 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
0.5 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.1 |
0.8 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 |
0.6 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 |
0.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.1 |
1.2 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 |
0.4 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 |
0.6 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.1 |
0.2 |
GO:0033128 |
negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 |
0.2 |
GO:0035617 |
stress granule disassembly(GO:0035617) |
| 0.0 |
0.3 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.5 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0002865 |
negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.0 |
0.6 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 |
0.4 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 |
0.3 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.5 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.6 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 |
0.3 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.3 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.9 |
GO:0006359 |
regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 |
0.2 |
GO:2001270 |
regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) |
| 0.0 |
0.1 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.0 |
GO:0045659 |
regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 |
0.3 |
GO:0051386 |
regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 |
0.1 |
GO:1902277 |
negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.0 |
0.7 |
GO:0009950 |
dorsal/ventral axis specification(GO:0009950) |
| 0.0 |
0.2 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.1 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 |
0.2 |
GO:1902410 |
mitotic cytokinetic process(GO:1902410) |
| 0.0 |
0.5 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.0 |
2.3 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.2 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 |
0.4 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.0 |
0.3 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.8 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.2 |
GO:0043922 |
negative regulation by host of viral transcription(GO:0043922) |
| 0.0 |
0.1 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.1 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.0 |
0.1 |
GO:0002667 |
lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 |
0.3 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.0 |
0.2 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.0 |
0.4 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.1 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 |
0.2 |
GO:0030238 |
male sex determination(GO:0030238) |
| 0.0 |
0.1 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0097190 |
apoptotic signaling pathway(GO:0097190) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.3 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.3 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.1 |
GO:0061484 |
hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 |
1.1 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.2 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.3 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.2 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 |
0.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.4 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.0 |
GO:0072356 |
chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 |
0.2 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.1 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |