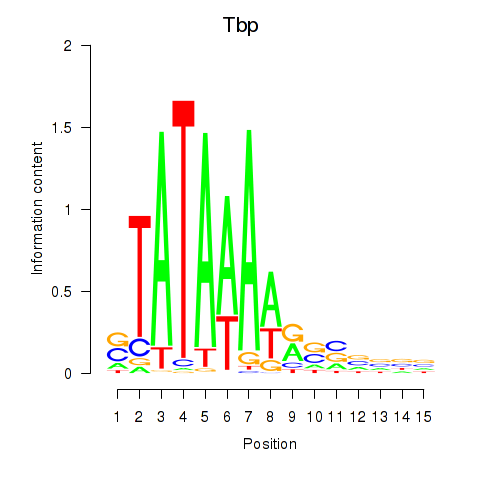
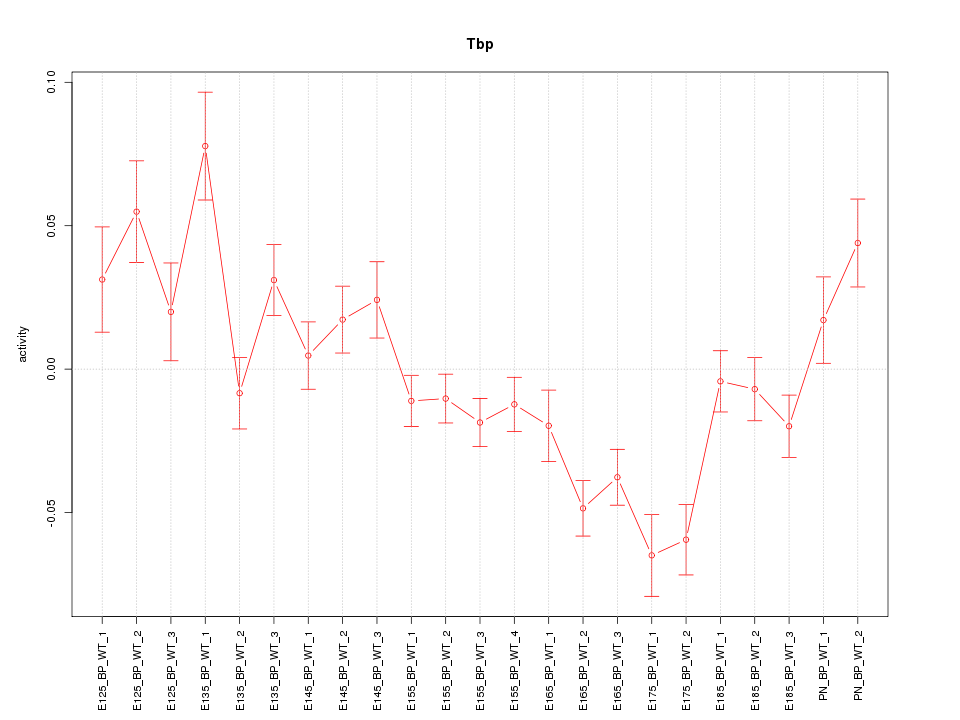
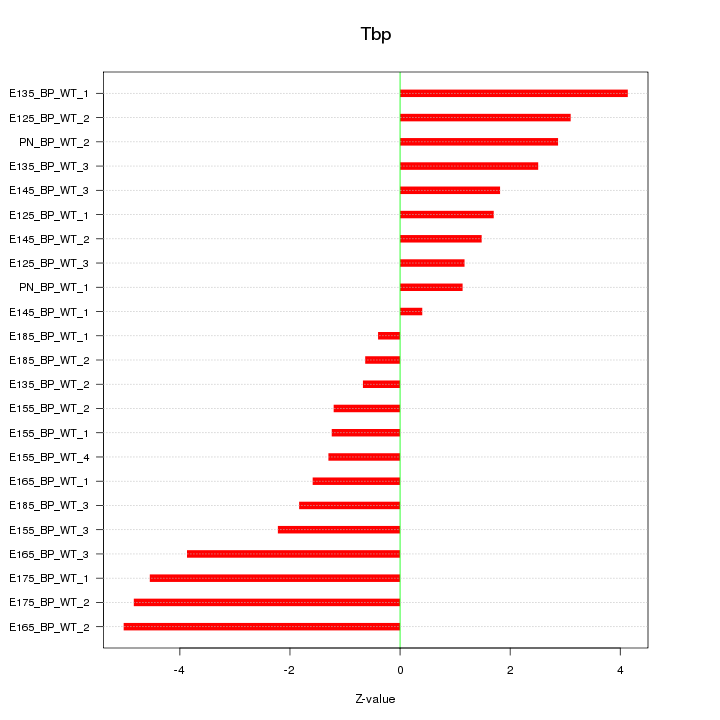
| 4.5 |
13.6 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 4.0 |
20.2 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.6 |
8.0 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 1.6 |
6.3 |
GO:0042414 |
epinephrine metabolic process(GO:0042414) |
| 1.4 |
18.1 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) positive regulation of collateral sprouting(GO:0048672) |
| 1.4 |
4.1 |
GO:0090285 |
negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 1.3 |
4.0 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) |
| 1.3 |
5.2 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 1.1 |
3.4 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 1.1 |
33.1 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.0 |
3.1 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 1.0 |
4.2 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.9 |
11.1 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.9 |
2.7 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.9 |
5.3 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.9 |
4.3 |
GO:1900147 |
Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.8 |
5.0 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.8 |
4.9 |
GO:0042535 |
positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.8 |
3.2 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.8 |
3.2 |
GO:0045091 |
regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.8 |
3.0 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.8 |
2.3 |
GO:0034124 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.7 |
2.2 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.7 |
5.1 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 0.7 |
2.2 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.7 |
4.7 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.7 |
2.0 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.6 |
4.8 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.6 |
4.0 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.6 |
2.8 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.5 |
2.7 |
GO:0010273 |
detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.5 |
3.7 |
GO:0016560 |
protein import into peroxisome matrix, docking(GO:0016560) |
| 0.5 |
5.1 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.5 |
6.5 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.4 |
1.3 |
GO:1900020 |
ovarian follicle rupture(GO:0001543) angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) establishment of blood-nerve barrier(GO:0008065) regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) regulation of membrane hyperpolarization(GO:1902630) |
| 0.4 |
7.7 |
GO:0000920 |
cell separation after cytokinesis(GO:0000920) |
| 0.4 |
1.3 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.4 |
1.3 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.4 |
1.7 |
GO:1903237 |
negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.4 |
4.1 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.4 |
1.2 |
GO:0010899 |
regulation of phosphatidylcholine catabolic process(GO:0010899) |
| 0.4 |
1.2 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) negative regulation of lymphocyte migration(GO:2000402) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 |
1.2 |
GO:0045876 |
positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.4 |
1.2 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.4 |
1.8 |
GO:0033087 |
negative regulation of immature T cell proliferation(GO:0033087) |
| 0.3 |
5.2 |
GO:0019430 |
removal of superoxide radicals(GO:0019430) |
| 0.3 |
1.3 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.3 |
3.6 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.3 |
0.9 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 |
0.9 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.3 |
1.8 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.3 |
1.1 |
GO:1902268 |
negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 |
0.8 |
GO:0044650 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) |
| 0.3 |
0.8 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) |
| 0.3 |
2.1 |
GO:0045842 |
positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.3 |
1.3 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.2 |
1.5 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.2 |
0.7 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.2 |
0.7 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 |
3.5 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 |
14.9 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.2 |
0.6 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.2 |
1.4 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.2 |
0.8 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.2 |
1.3 |
GO:1902416 |
positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.2 |
1.3 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 |
0.6 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.2 |
0.9 |
GO:1903026 |
negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 |
0.5 |
GO:0042713 |
negative regulation of urine volume(GO:0035811) sperm ejaculation(GO:0042713) negative regulation of gastric acid secretion(GO:0060455) |
| 0.2 |
0.5 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 |
2.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.1 |
0.4 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
2.2 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 |
1.9 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
1.5 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
2.5 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 |
1.1 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.1 |
0.5 |
GO:1903243 |
negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 |
1.6 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
2.4 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.1 |
1.9 |
GO:0031987 |
locomotion involved in locomotory behavior(GO:0031987) |
| 0.1 |
0.8 |
GO:0046688 |
response to copper ion(GO:0046688) |
| 0.1 |
0.3 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 |
1.0 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
1.2 |
GO:0071260 |
cellular response to mechanical stimulus(GO:0071260) |
| 0.1 |
2.1 |
GO:0071398 |
cellular response to fatty acid(GO:0071398) |
| 0.1 |
5.9 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 |
0.5 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 |
2.2 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.1 |
1.4 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 |
0.9 |
GO:0002548 |
monocyte chemotaxis(GO:0002548) |
| 0.1 |
2.3 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
0.5 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 0.1 |
0.7 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.1 |
1.7 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.1 |
0.6 |
GO:0000712 |
resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 |
0.2 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
7.2 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.1 |
0.2 |
GO:0060854 |
patterning of lymph vessels(GO:0060854) |
| 0.1 |
3.4 |
GO:0022904 |
respiratory electron transport chain(GO:0022904) |
| 0.1 |
1.1 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
1.6 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
0.2 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.1 |
0.5 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 0.1 |
0.2 |
GO:1903225 |
regulation of endodermal cell fate specification(GO:0042663) negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 |
1.7 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.1 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 |
3.3 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.0 |
0.7 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.0 |
0.5 |
GO:0060965 |
negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 |
0.6 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.0 |
0.6 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 |
1.5 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
1.0 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.2 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.4 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 |
0.4 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 |
1.2 |
GO:0008286 |
insulin receptor signaling pathway(GO:0008286) |
| 0.0 |
0.8 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.9 |
GO:0042752 |
regulation of circadian rhythm(GO:0042752) |
| 0.0 |
1.0 |
GO:0051225 |
spindle assembly(GO:0051225) |
| 0.0 |
0.6 |
GO:0043010 |
camera-type eye development(GO:0043010) |