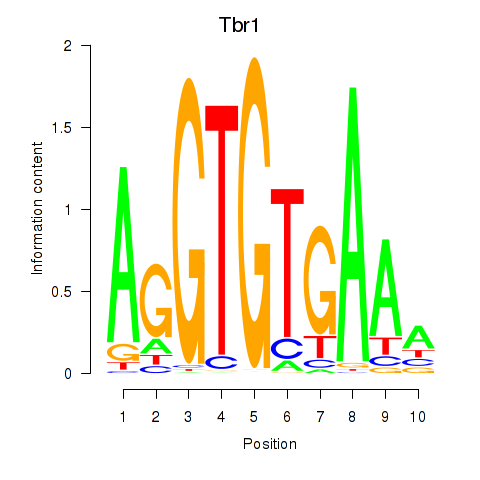
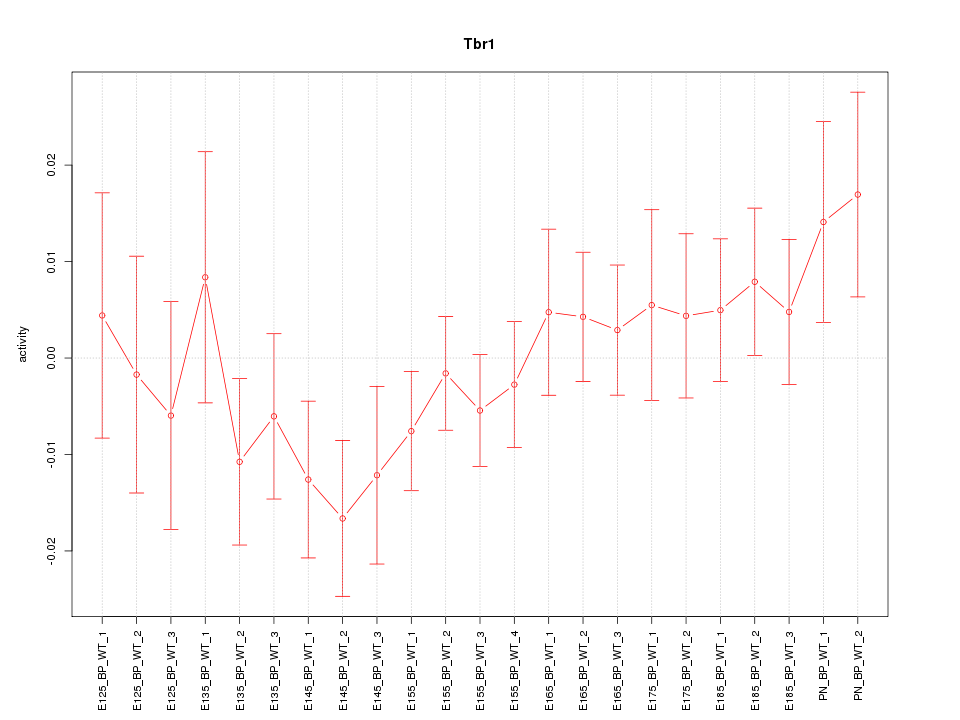
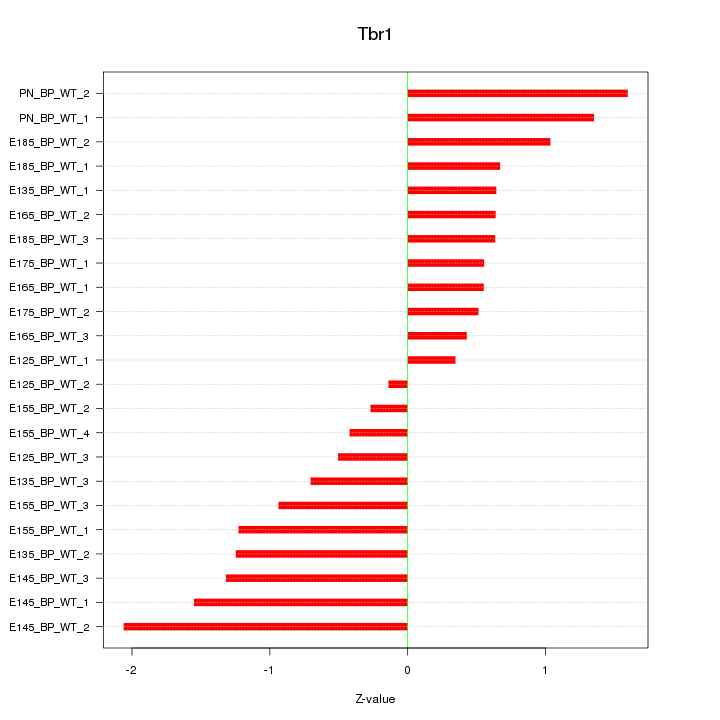
| 1.6 |
4.8 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.7 |
2.2 |
GO:0048818 |
positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.6 |
1.8 |
GO:0030421 |
defecation(GO:0030421) |
| 0.6 |
1.8 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 0.5 |
3.4 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.4 |
1.1 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 |
1.8 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.4 |
0.7 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.3 |
1.0 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.3 |
1.0 |
GO:1901628 |
positive regulation of postsynaptic membrane organization(GO:1901628) |
| 0.3 |
1.9 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 |
0.9 |
GO:1904706 |
negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.3 |
0.9 |
GO:0003245 |
cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 |
1.1 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.3 |
0.8 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.3 |
0.8 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.3 |
1.1 |
GO:2000048 |
negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.3 |
0.8 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.3 |
1.5 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.3 |
1.3 |
GO:1901679 |
nucleotide transmembrane transport(GO:1901679) |
| 0.2 |
0.7 |
GO:0070973 |
protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 |
0.9 |
GO:0032610 |
interleukin-1 alpha production(GO:0032610) |
| 0.2 |
0.9 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
1.1 |
GO:2001204 |
regulation of osteoclast development(GO:2001204) |
| 0.2 |
1.1 |
GO:2000483 |
negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.2 |
0.6 |
GO:1990481 |
snoRNA guided rRNA pseudouridine synthesis(GO:0000454) snRNA pseudouridine synthesis(GO:0031120) mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 |
1.3 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 |
0.6 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.2 |
0.7 |
GO:0051593 |
response to folic acid(GO:0051593) |
| 0.2 |
0.7 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.2 |
1.0 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.2 |
0.5 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.2 |
0.5 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.2 |
0.5 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 |
3.2 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.2 |
1.2 |
GO:0045591 |
positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.2 |
0.8 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.2 |
0.6 |
GO:0035934 |
corticosterone secretion(GO:0035934) |
| 0.2 |
0.8 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.1 |
0.7 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.7 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 |
0.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.1 |
0.4 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
0.7 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 |
0.8 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.1 |
0.6 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.6 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 |
0.4 |
GO:0045204 |
MAPK export from nucleus(GO:0045204) |
| 0.1 |
0.4 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 |
2.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.7 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
2.1 |
GO:0035994 |
response to muscle stretch(GO:0035994) |
| 0.1 |
0.5 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.5 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.1 |
1.6 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
0.4 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.1 |
0.4 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 |
0.6 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 |
0.5 |
GO:0007066 |
female meiosis sister chromatid cohesion(GO:0007066) chiasma assembly(GO:0051026) |
| 0.1 |
0.4 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.6 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.1 |
1.1 |
GO:0009992 |
cellular water homeostasis(GO:0009992) |
| 0.1 |
0.3 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.5 |
GO:0006668 |
sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.1 |
1.4 |
GO:0042415 |
norepinephrine metabolic process(GO:0042415) |
| 0.1 |
0.8 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.1 |
0.4 |
GO:0071449 |
cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 |
0.3 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 |
0.3 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.1 |
0.6 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.1 |
0.9 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.1 |
0.3 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.8 |
GO:0035735 |
intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 |
0.8 |
GO:0042730 |
fibrinolysis(GO:0042730) |
| 0.1 |
2.5 |
GO:0032757 |
positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 |
0.5 |
GO:0060648 |
mammary gland bud morphogenesis(GO:0060648) |
| 0.1 |
4.7 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.7 |
GO:0009181 |
purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 |
0.4 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.1 |
0.3 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.1 |
0.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 |
1.4 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 |
0.3 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
0.5 |
GO:0006172 |
ADP biosynthetic process(GO:0006172) |
| 0.1 |
0.2 |
GO:0015867 |
ATP transport(GO:0015867) |
| 0.1 |
0.3 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.5 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 |
1.0 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.1 |
GO:0070366 |
regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 |
0.5 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.7 |
GO:0035562 |
negative regulation of chromatin binding(GO:0035562) |
| 0.1 |
0.4 |
GO:0048254 |
snoRNA localization(GO:0048254) |
| 0.1 |
0.3 |
GO:0071267 |
amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 |
0.3 |
GO:2000427 |
eosinophil chemotaxis(GO:0048245) T cell extravasation(GO:0072683) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 |
0.3 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 |
0.2 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 |
0.4 |
GO:0045605 |
negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 |
0.2 |
GO:0032497 |
detection of lipopolysaccharide(GO:0032497) |
| 0.1 |
0.6 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
3.0 |
GO:0046676 |
negative regulation of insulin secretion(GO:0046676) |
| 0.1 |
0.2 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.1 |
0.5 |
GO:2000194 |
regulation of female gonad development(GO:2000194) |
| 0.1 |
0.3 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.2 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.1 |
0.1 |
GO:2000347 |
positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.1 |
0.5 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.3 |
GO:0050747 |
positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.1 |
0.3 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.1 |
0.7 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.1 |
0.7 |
GO:0072010 |
renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.1 |
0.2 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) resolution of recombination intermediates(GO:0071139) |
| 0.1 |
0.5 |
GO:0044130 |
negative regulation of growth of symbiont in host(GO:0044130) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.1 |
0.3 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.1 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.1 |
0.4 |
GO:0043504 |
mitochondrial DNA repair(GO:0043504) |
| 0.1 |
0.5 |
GO:0043402 |
glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 |
0.2 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.1 |
0.4 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.5 |
GO:0006544 |
glycine metabolic process(GO:0006544) |
| 0.1 |
0.4 |
GO:0044791 |
modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 |
0.6 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 |
0.3 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 |
0.1 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.0 |
0.3 |
GO:0002834 |
immune response to tumor cell(GO:0002418) regulation of response to tumor cell(GO:0002834) regulation of immune response to tumor cell(GO:0002837) |
| 0.0 |
0.5 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.9 |
GO:2000095 |
regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 |
0.1 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.0 |
0.3 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
1.1 |
GO:0030866 |
cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 |
1.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.2 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.1 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 |
0.2 |
GO:0006478 |
peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 |
0.1 |
GO:0006667 |
sphinganine metabolic process(GO:0006667) |
| 0.0 |
0.6 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.2 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 |
0.8 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
0.1 |
GO:0031652 |
positive regulation of heat generation(GO:0031652) |
| 0.0 |
0.2 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 |
0.4 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 |
0.2 |
GO:1904715 |
negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 |
0.5 |
GO:1901897 |
regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 |
0.3 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.5 |
GO:2001014 |
regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 |
0.6 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.4 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
1.3 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.0 |
0.3 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 |
1.0 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
0.6 |
GO:0048642 |
negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 |
0.1 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 |
0.4 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 |
0.7 |
GO:0044253 |
positive regulation of collagen metabolic process(GO:0010714) positive regulation of collagen biosynthetic process(GO:0032967) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 |
0.2 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 |
0.3 |
GO:0044331 |
cell-cell adhesion mediated by cadherin(GO:0044331) |
| 0.0 |
0.5 |
GO:0060049 |
regulation of protein glycosylation(GO:0060049) |
| 0.0 |
0.2 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.0 |
0.3 |
GO:0001672 |
regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 |
0.2 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 |
0.4 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 |
0.4 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.2 |
GO:0098734 |
protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.1 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 |
0.2 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.2 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
0.2 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.1 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.2 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.2 |
GO:0001980 |
regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 |
1.2 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.4 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.0 |
1.2 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 |
0.2 |
GO:1990928 |
response to amino acid starvation(GO:1990928) |
| 0.0 |
0.4 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.6 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.2 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.6 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
1.4 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.1 |
GO:0038108 |
negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 |
0.1 |
GO:2000482 |
interleukin-8 secretion(GO:0072606) regulation of interleukin-8 secretion(GO:2000482) |
| 0.0 |
0.3 |
GO:1904031 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 |
1.3 |
GO:0048821 |
erythrocyte development(GO:0048821) |
| 0.0 |
0.6 |
GO:0046325 |
negative regulation of glucose import(GO:0046325) |
| 0.0 |
1.4 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.0 |
0.1 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 |
0.3 |
GO:2000406 |
positive regulation of T cell migration(GO:2000406) |
| 0.0 |
0.3 |
GO:0050858 |
negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.7 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
1.3 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.0 |
0.9 |
GO:0070306 |
lens fiber cell differentiation(GO:0070306) |
| 0.0 |
0.2 |
GO:0060575 |
intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 |
0.4 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
0.2 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.0 |
0.1 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0070995 |
NADPH oxidation(GO:0070995) |
| 0.0 |
0.8 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.1 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.4 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.3 |
GO:0042573 |
retinoic acid metabolic process(GO:0042573) |
| 0.0 |
0.2 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.1 |
GO:0010793 |
regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.0 |
0.8 |
GO:0035196 |
production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 |
0.1 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 |
0.1 |
GO:0060431 |
primary lung bud formation(GO:0060431) |
| 0.0 |
0.1 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
0.1 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 |
0.2 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.4 |
GO:0040036 |
regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 |
0.4 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.1 |
GO:1903984 |
regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.1 |
GO:0097009 |
energy homeostasis(GO:0097009) |
| 0.0 |
0.6 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.0 |
0.3 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 |
0.1 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.0 |
GO:1903722 |
regulation of centriole elongation(GO:1903722) |
| 0.0 |
0.1 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.1 |
GO:0010826 |
negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.2 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.0 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.0 |
0.3 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.0 |
0.7 |
GO:0051453 |
regulation of intracellular pH(GO:0051453) |
| 0.0 |
0.2 |
GO:0032460 |
negative regulation of protein oligomerization(GO:0032460) negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.2 |
GO:0051457 |
maintenance of protein location in nucleus(GO:0051457) |
| 0.0 |
0.2 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 |
0.2 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 |
0.1 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.0 |
0.4 |
GO:0008156 |
negative regulation of DNA replication(GO:0008156) |
| 0.0 |
0.1 |
GO:0043313 |
regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 |
0.1 |
GO:0006105 |
succinate metabolic process(GO:0006105) |
| 0.0 |
0.2 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.7 |
GO:0030500 |
regulation of bone mineralization(GO:0030500) |
| 0.0 |
0.1 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
1.0 |
GO:0042147 |
retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 |
0.1 |
GO:0033147 |
negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 |
0.1 |
GO:0006968 |
cellular defense response(GO:0006968) |
| 0.0 |
0.0 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 |
0.3 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.0 |
0.3 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.2 |
GO:0010453 |
regulation of cell fate commitment(GO:0010453) |
| 0.0 |
0.6 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.0 |
0.3 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
1.0 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.1 |
GO:0032462 |
regulation of protein homooligomerization(GO:0032462) |
| 0.0 |
0.1 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
0.1 |
GO:0006532 |
aspartate biosynthetic process(GO:0006532) |
| 0.0 |
0.3 |
GO:0045620 |
negative regulation of lymphocyte differentiation(GO:0045620) |
| 0.0 |
0.5 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.2 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.0 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 |
0.1 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
0.1 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.3 |
GO:0015758 |
glucose transport(GO:0015758) |