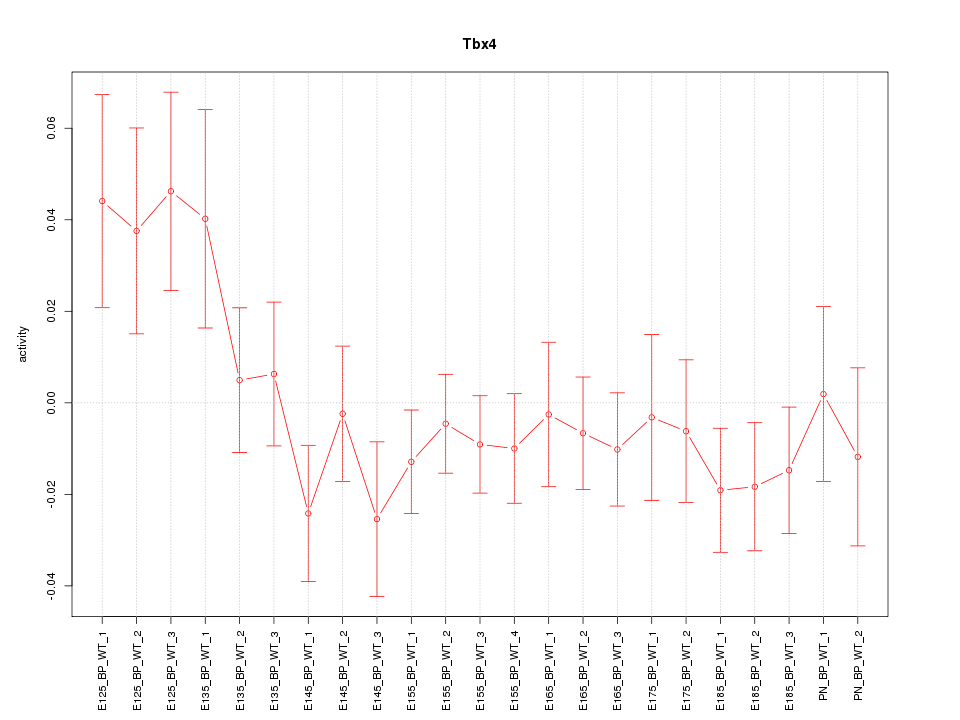
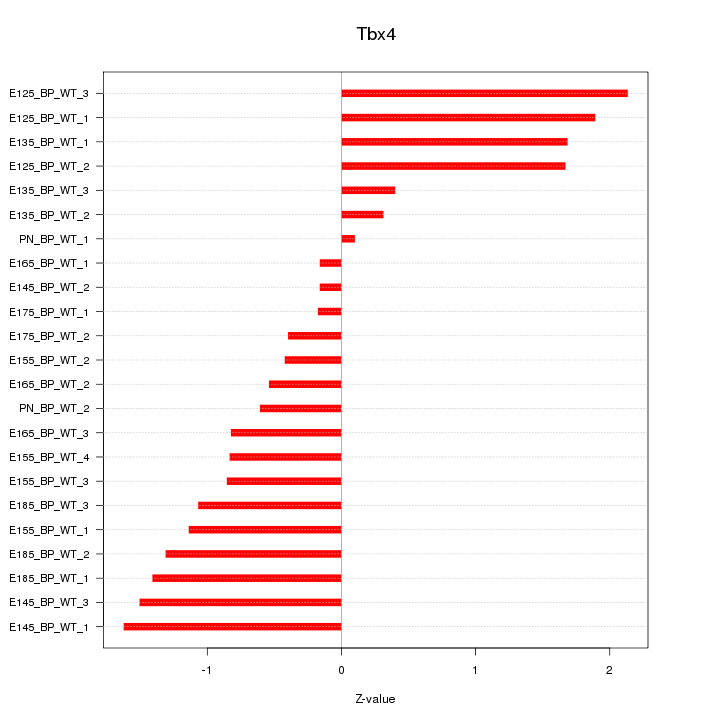
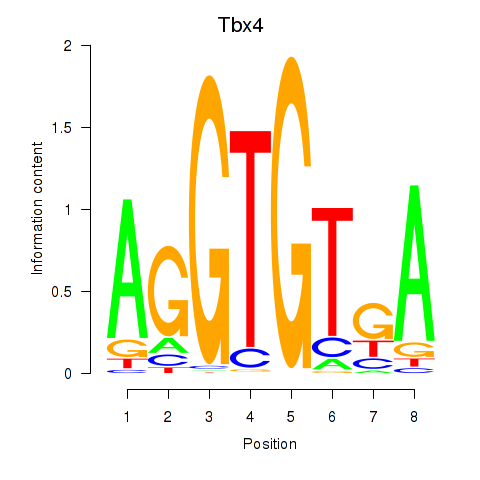
Motif ID: Tbx4
Z-value: 1.109
Transcription factors associated with Tbx4:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tbx4 | ENSMUSG00000000094.6 | Tbx4 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 1.9 | 9.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 1.7 | 6.6 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 1.2 | 3.6 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.9 | 2.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.8 | 4.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.8 | 7.4 | GO:0071493 | cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.7 | 9.7 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.7 | 2.1 | GO:0097623 | potassium ion export across plasma membrane(GO:0097623) |
| 0.5 | 2.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.5 | 4.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.5 | 5.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 1.4 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 | 1.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 | 3.3 | GO:0044027 | hypermethylation of CpG island(GO:0044027) |
| 0.3 | 1.2 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 | 1.7 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 1.4 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 1.0 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 3.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.1 | 1.8 | GO:0097067 | negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 2.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 0.6 | GO:0071638 | cellular triglyceride homeostasis(GO:0035356) adiponectin secretion(GO:0070162) regulation of adiponectin secretion(GO:0070163) negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.5 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 2.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.6 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.6 | GO:0019348 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) dolichol metabolic process(GO:0019348) |
| 0.1 | 2.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 0.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.7 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 | 0.8 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 8.8 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.1 | 2.9 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.3 | GO:2000676 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.3 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.6 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 5.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 | 0.9 | GO:0071385 | cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 1.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 2.3 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.5 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.8 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.3 | 1.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.3 | 2.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 9.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.3 | GO:0001741 | XY body(GO:0001741) |
| 0.1 | 9.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 4.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.6 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 4.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.0 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.6 | 1.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.5 | 7.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 8.6 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.4 | 3.3 | GO:0051718 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) DNA (cytosine-5-)-methyltransferase activity, acting on CpG substrates(GO:0051718) |
| 0.4 | 2.7 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.3 | 9.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 4.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.2 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 0.6 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 0.2 | 0.6 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 4.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 3.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 5.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 1.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.9 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 2.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.6 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 5.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 22.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 2.6 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) NADPH:sulfur oxidoreductase activity(GO:0043914) epoxyqueuosine reductase activity(GO:0052693) |