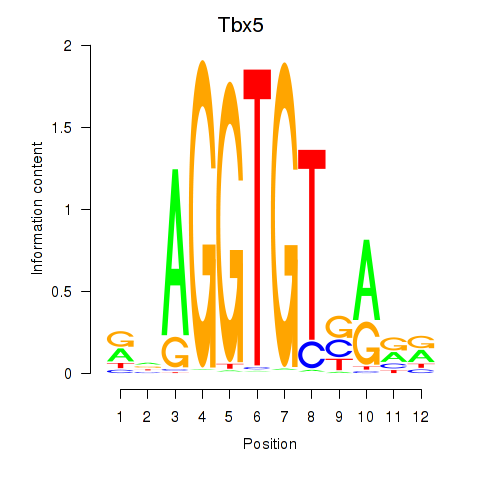
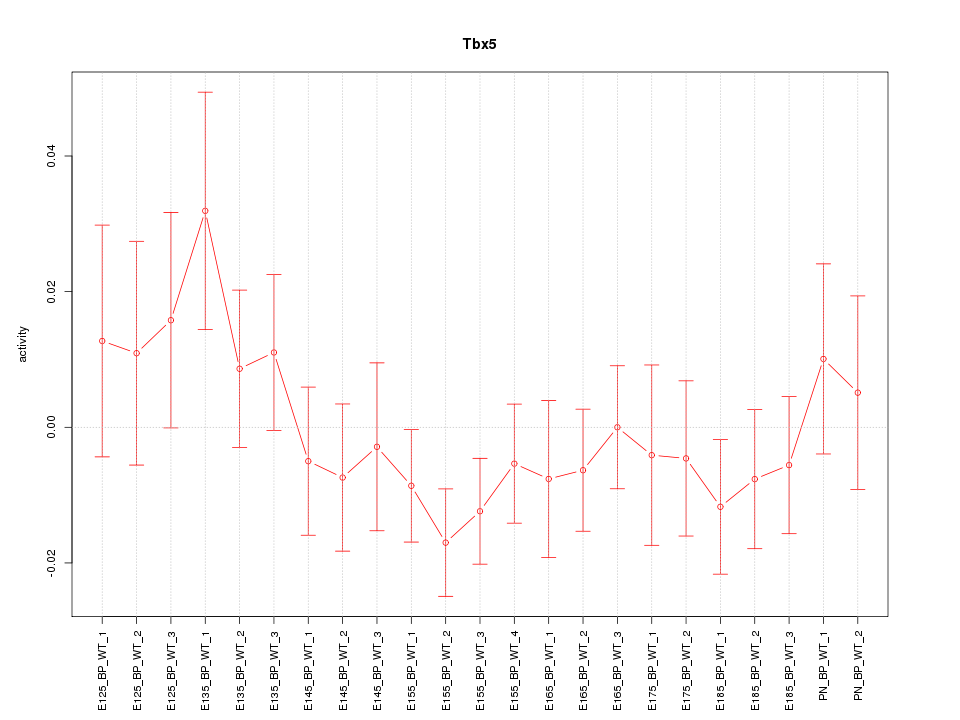
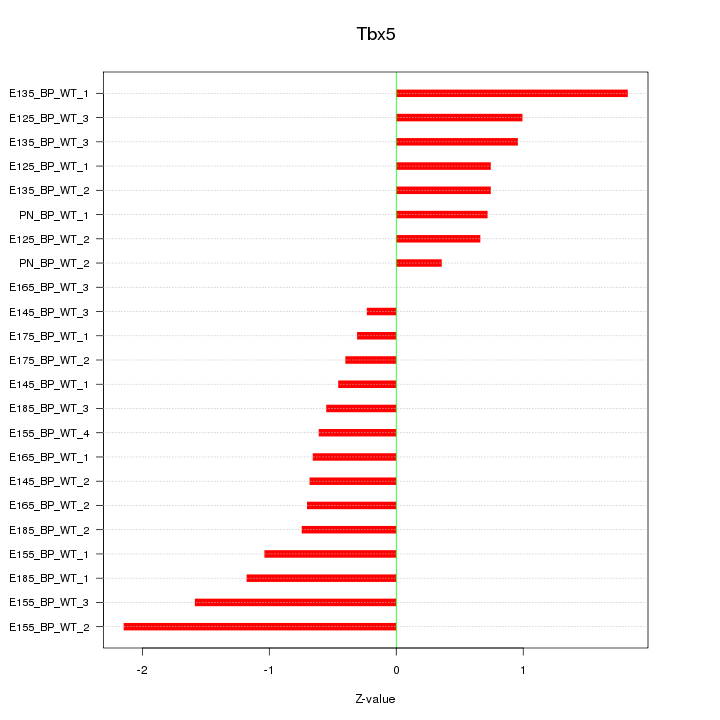
| 1.2 |
6.2 |
GO:0035022 |
positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.8 |
3.8 |
GO:0035262 |
gonad morphogenesis(GO:0035262) |
| 0.6 |
2.6 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 |
1.8 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.6 |
1.7 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.5 |
1.5 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.4 |
2.2 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.4 |
1.5 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.4 |
1.1 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.4 |
1.1 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.3 |
1.7 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.3 |
1.0 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.3 |
2.2 |
GO:0043951 |
negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.3 |
0.9 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) |
| 0.3 |
3.4 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.2 |
1.2 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
2.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.2 |
1.8 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 |
1.2 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.2 |
1.4 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 |
0.8 |
GO:0071139 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) resolution of recombination intermediates(GO:0071139) |
| 0.2 |
0.6 |
GO:0007403 |
glial cell fate determination(GO:0007403) |
| 0.2 |
0.4 |
GO:0016264 |
gap junction assembly(GO:0016264) |
| 0.2 |
0.6 |
GO:0040009 |
regulation of growth rate(GO:0040009) |
| 0.2 |
0.5 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.2 |
0.5 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.2 |
1.2 |
GO:0031508 |
pericentric heterochromatin assembly(GO:0031508) |
| 0.2 |
1.8 |
GO:0045606 |
positive regulation of epidermal cell differentiation(GO:0045606) |
| 0.2 |
0.8 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 |
0.6 |
GO:0048304 |
positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 |
0.5 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 |
0.4 |
GO:0003104 |
positive regulation of glomerular filtration(GO:0003104) cell communication by chemical coupling(GO:0010643) |
| 0.1 |
1.3 |
GO:0014824 |
artery smooth muscle contraction(GO:0014824) |
| 0.1 |
0.6 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.1 |
0.4 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 |
1.0 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.1 |
1.9 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
1.6 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.5 |
GO:1902608 |
regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 |
0.4 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.1 |
0.5 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.1 |
2.2 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.1 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.1 |
1.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.7 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 |
0.3 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.1 |
0.4 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 |
0.9 |
GO:1904153 |
negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.1 |
1.0 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.1 |
0.2 |
GO:0042822 |
vitamin B6 metabolic process(GO:0042816) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 |
0.5 |
GO:0043415 |
positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 |
0.3 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.1 |
0.9 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
1.4 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 |
1.4 |
GO:0060236 |
regulation of mitotic spindle organization(GO:0060236) |
| 0.1 |
0.2 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
0.2 |
GO:0060217 |
positive regulation of chromatin assembly or disassembly(GO:0045799) hemangioblast cell differentiation(GO:0060217) |
| 0.1 |
1.2 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.6 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.1 |
1.7 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.1 |
0.3 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.4 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
0.4 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
1.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 |
0.3 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.1 |
0.9 |
GO:0060670 |
branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 |
0.6 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.3 |
GO:0010025 |
wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 |
0.3 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.4 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.0 |
0.1 |
GO:0097089 |
methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 |
1.7 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.5 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.0 |
0.8 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 |
0.3 |
GO:0090195 |
positive regulation of T-helper 2 cell differentiation(GO:0045630) chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 |
0.6 |
GO:2000193 |
positive regulation of fatty acid transport(GO:2000193) |
| 0.0 |
0.1 |
GO:0051835 |
positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 |
0.1 |
GO:0045014 |
carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 |
0.3 |
GO:0042523 |
positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 |
0.9 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.1 |
GO:0009812 |
flavonoid metabolic process(GO:0009812) flavonoid biosynthetic process(GO:0009813) flavonoid glucuronidation(GO:0052696) |
| 0.0 |
0.1 |
GO:2000334 |
lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 |
0.7 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
1.0 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.3 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.0 |
0.3 |
GO:0032780 |
negative regulation of ATPase activity(GO:0032780) |
| 0.0 |
0.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
2.4 |
GO:0031638 |
zymogen activation(GO:0031638) |
| 0.0 |
0.2 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.2 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.0 |
0.2 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.0 |
0.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
0.3 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.0 |
0.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.0 |
0.9 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.1 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.8 |
GO:0035774 |
positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 |
2.9 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.8 |
GO:0007050 |
cell cycle arrest(GO:0007050) |
| 0.0 |
0.7 |
GO:0045103 |
intermediate filament-based process(GO:0045103) |
| 0.0 |
0.1 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.5 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.0 |
0.4 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.5 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.3 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.0 |
0.4 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
0.7 |
GO:0032465 |
regulation of cytokinesis(GO:0032465) |
| 0.0 |
0.2 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.3 |
GO:0032958 |
inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 |
0.1 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:0007602 |
phototransduction(GO:0007602) |
| 0.0 |
0.0 |
GO:1900060 |
negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 |
0.2 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.4 |
GO:0034766 |
negative regulation of ion transmembrane transport(GO:0034766) |
| 0.0 |
1.1 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.0 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 |
0.4 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.7 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.4 |
GO:0070534 |
protein K63-linked ubiquitination(GO:0070534) |
| 0.0 |
0.3 |
GO:1902850 |
mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 |
0.2 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.1 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |