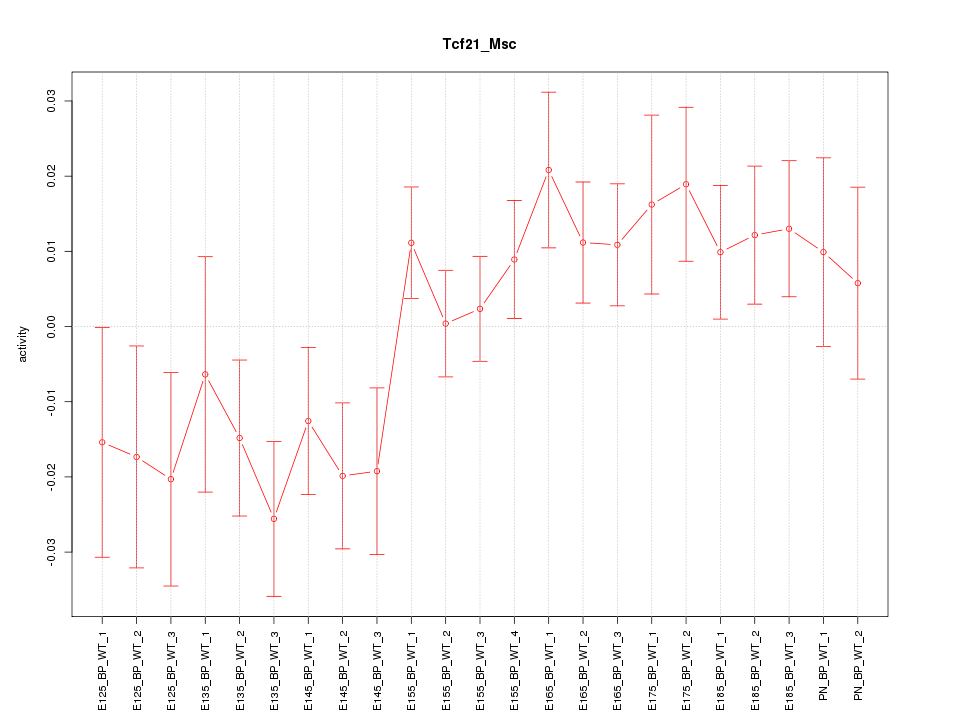
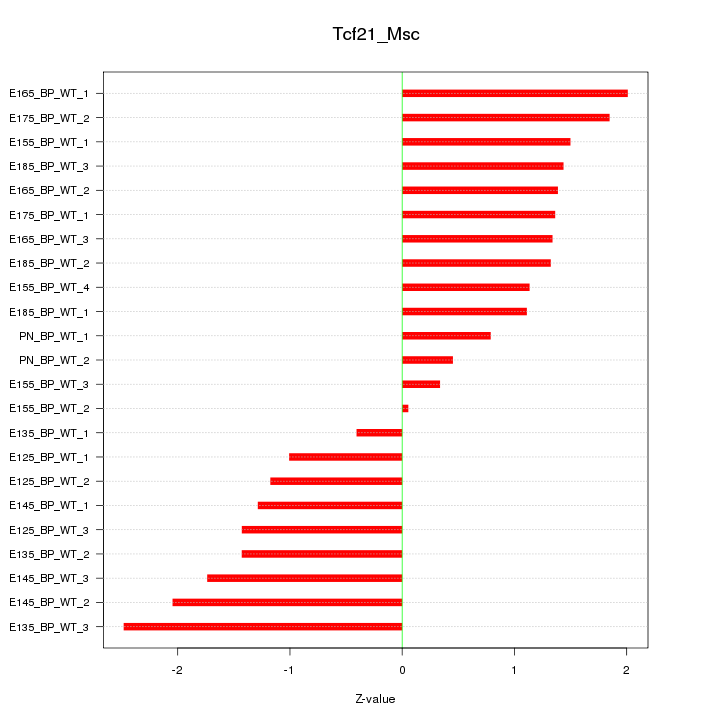
Motif ID: Tcf21_Msc
Z-value: 1.387
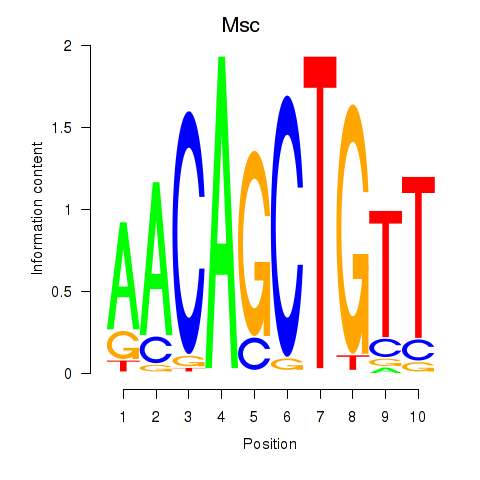
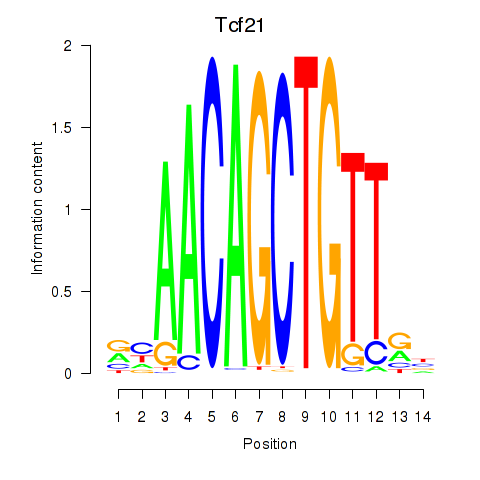
Transcription factors associated with Tcf21_Msc:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Msc | ENSMUSG00000025930.5 | Msc |
| Tcf21 | ENSMUSG00000045680.7 | Tcf21 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 9.4 | GO:0038095 | positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.9 | 21.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.3 | 4.0 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 1.0 | 6.3 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.0 | 4.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.7 | 4.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.7 | 7.0 | GO:0007379 | segment specification(GO:0007379) |
| 0.7 | 3.4 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.6 | 3.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.5 | 1.6 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.5 | 1.6 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.4 | 1.3 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 1.2 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.4 | 6.6 | GO:1901629 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) regulation of presynaptic membrane organization(GO:1901629) |
| 0.4 | 1.6 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.4 | 1.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.4 | 2.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 3.4 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.4 | 1.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.4 | 3.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.4 | 1.4 | GO:0099624 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.3 | 1.0 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.3 | 1.0 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.3 | 0.3 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.3 | 2.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.3 | 0.9 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 | 0.3 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.3 | 1.9 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 | 1.1 | GO:0042636 | striatal medium spiny neuron differentiation(GO:0021773) negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.3 | 1.9 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 0.8 | GO:1904049 | regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.3 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 0.5 | GO:0060467 | negative regulation of fertilization(GO:0060467) |
| 0.3 | 2.5 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.3 | 2.0 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 2.0 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 1.2 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 2.9 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 | 0.7 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 0.5 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) ureter morphogenesis(GO:0072197) |
| 0.2 | 2.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.9 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 0.7 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.2 | 1.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.2 | 0.6 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.2 | 3.9 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 0.6 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.2 | 1.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 | 0.6 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.9 | GO:0030432 | peristalsis(GO:0030432) |
| 0.2 | 2.0 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.2 | 0.9 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.2 | 0.7 | GO:0032439 | endosome localization(GO:0032439) |
| 0.2 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.2 | 1.6 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.2 | 2.8 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 0.5 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.2 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 1.0 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.2 | 2.0 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.6 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.4 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.1 | 0.4 | GO:0039521 | suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 1.2 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 0.4 | GO:1901675 | negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.1 | 0.4 | GO:0019478 | D-amino acid catabolic process(GO:0019478) |
| 0.1 | 0.5 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.1 | 0.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.1 | 0.9 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 4.0 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.4 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.1 | 2.2 | GO:0051043 | regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.6 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 3.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.2 | GO:2000409 | positive regulation of T cell extravasation(GO:2000409) |
| 0.1 | 3.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.3 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.2 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.5 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.5 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.7 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.9 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.1 | 1.8 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.3 | GO:0046168 | glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 1.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 1.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.3 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.1 | 0.3 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.1 | 0.3 | GO:0003032 | detection of oxygen(GO:0003032) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 0.1 | 0.3 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 1.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.1 | 1.2 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 2.2 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 2.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 0.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.1 | 1.6 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.5 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.1 | 8.2 | GO:0051592 | response to calcium ion(GO:0051592) |
| 0.1 | 0.7 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 0.3 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.8 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.1 | 0.9 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.1 | 0.8 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.1 | 0.6 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.7 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 0.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.3 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.6 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.2 | GO:0070589 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.4 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0050904 | diapedesis(GO:0050904) |
| 0.1 | 0.6 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.1 | 0.3 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.2 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0045072 | regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.5 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.9 | GO:0001523 | retinoid metabolic process(GO:0001523) |
| 0.0 | 0.9 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.2 | GO:0046210 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.0 | 0.4 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.4 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.0 | 0.2 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.8 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 1.0 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.3 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.5 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 1.5 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 3.8 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.1 | GO:0090155 | negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.0 | 2.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.8 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.3 | GO:0050965 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.1 | GO:0031052 | programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.0 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 1.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 1.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.3 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.4 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.4 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.4 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.3 | GO:0030501 | positive regulation of bone mineralization(GO:0030501) |
| 0.0 | 0.3 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0006721 | terpenoid metabolic process(GO:0006721) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 1.0 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 1.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.1 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.0 | 0.1 | GO:0007625 | grooming behavior(GO:0007625) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.7 | 2.2 | GO:0071438 | integrin alpha3-beta1 complex(GO:0034667) invadopodium membrane(GO:0071438) |
| 0.4 | 1.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.4 | 1.6 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.4 | 1.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.4 | 2.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.3 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 0.3 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.3 | 0.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 0.9 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 1.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 9.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 8.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.2 | 1.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 2.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 3.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.2 | 0.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.9 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.8 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.1 | 1.0 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.4 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 6.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 1.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.1 | 1.4 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.5 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.1 | 0.7 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.4 | GO:0001651 | dense fibrillar component(GO:0001651) box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.1 | 1.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 2.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 2.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.2 | GO:0031502 | dolichyl-phosphate-mannose-protein mannosyltransferase complex(GO:0031502) |
| 0.1 | 1.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.4 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 1.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.3 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.0 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 2.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 14.2 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 5.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 8.2 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.8 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 3.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.8 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.2 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 21.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.0 | 3.1 | GO:0005534 | galactose binding(GO:0005534) |
| 1.0 | 4.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.8 | 6.3 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.7 | 2.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.7 | 3.4 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.6 | 9.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.5 | 1.5 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.5 | 4.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.5 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.4 | 1.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.4 | 4.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 1.8 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.4 | 3.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.3 | 3.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.3 | 0.9 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.3 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.3 | 6.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.2 | 3.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.2 | 1.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.2 | 1.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.6 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.6 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.2 | 1.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.9 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.2 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 0.8 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.1 | 0.6 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.1 | 1.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 0.4 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 1.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 4.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 1.3 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 2.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 1.2 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 0.4 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.6 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.4 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.1 | 0.8 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 2.2 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.1 | 1.0 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 1.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 2.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 4.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.4 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) aspartate oxidase activity(GO:0015922) |
| 0.1 | 0.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.7 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 1.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.7 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.1 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.4 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.1 | 0.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 2.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.5 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.7 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.1 | 0.3 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.2 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 0.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.5 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.8 | GO:1901505 | carbohydrate derivative transporter activity(GO:1901505) |
| 0.0 | 0.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.0 | 1.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 6.5 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.4 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.3 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 1.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.5 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 3.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 1.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.7 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.0 | GO:0004376 | alpha-1,2-mannosyltransferase activity(GO:0000026) glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.6 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 3.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.0 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.4 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 0.0 | 0.5 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |