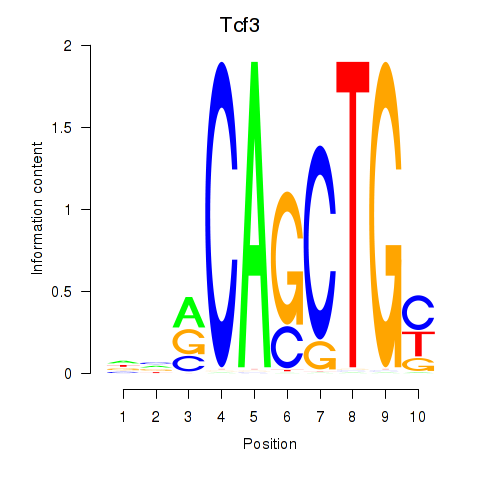
| 0.5 |
2.3 |
GO:0007386 |
compartment pattern specification(GO:0007386) |
| 0.3 |
0.9 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
0.7 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.2 |
0.6 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.2 |
0.8 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.2 |
0.6 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.2 |
0.9 |
GO:0071205 |
clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.2 |
1.3 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
0.5 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.2 |
0.7 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.2 |
0.5 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.2 |
0.5 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.1 |
0.6 |
GO:0032439 |
endosome localization(GO:0032439) |
| 0.1 |
0.4 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
0.3 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
0.4 |
GO:0016332 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 |
1.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 |
1.0 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.2 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.1 |
0.3 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.1 |
0.7 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
1.3 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
1.3 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 |
0.3 |
GO:0086017 |
Purkinje myocyte action potential(GO:0086017) |
| 0.1 |
0.8 |
GO:0001977 |
renal system process involved in regulation of blood volume(GO:0001977) |
| 0.1 |
0.1 |
GO:2000040 |
regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 |
0.7 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 |
0.6 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 |
0.2 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.1 |
0.5 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 |
1.1 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.3 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.2 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 0.1 |
0.6 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 |
0.6 |
GO:0021678 |
third ventricle development(GO:0021678) |
| 0.1 |
0.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.2 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.1 |
0.2 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.1 |
0.5 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.1 |
0.7 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 |
0.2 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 0.1 |
0.2 |
GO:2000832 |
protein-chromophore linkage(GO:0018298) negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.6 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.1 |
0.1 |
GO:0035754 |
B cell chemotaxis(GO:0035754) |
| 0.1 |
0.2 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 |
0.2 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 |
0.2 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
1.7 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.1 |
GO:1904879 |
positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 |
0.0 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.0 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.1 |
GO:2001137 |
positive regulation of endocytic recycling(GO:2001137) |
| 0.0 |
0.3 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 |
0.4 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 |
1.2 |
GO:0010719 |
negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 |
0.7 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 |
0.1 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 |
0.4 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.0 |
0.1 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.0 |
0.6 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.3 |
GO:0034351 |
negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 |
0.2 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 |
0.1 |
GO:1990034 |
calcium ion export from cell(GO:1990034) calcium ion import into cell(GO:1990035) |
| 0.0 |
0.4 |
GO:0042534 |
tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 |
0.1 |
GO:0001803 |
type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 |
0.1 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) dorsal root ganglion development(GO:1990791) |
| 0.0 |
0.3 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.0 |
0.2 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.0 |
GO:0042321 |
negative regulation of circadian sleep/wake cycle, sleep(GO:0042321) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 0.0 |
0.4 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.5 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 |
0.5 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
1.1 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.0 |
0.4 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
0.3 |
GO:0002693 |
positive regulation of cellular extravasation(GO:0002693) |
| 0.0 |
0.3 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.1 |
GO:0002315 |
marginal zone B cell differentiation(GO:0002315) |
| 0.0 |
0.2 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 |
0.2 |
GO:0061032 |
visceral serous pericardium development(GO:0061032) |
| 0.0 |
0.1 |
GO:0035574 |
histone H4-K20 demethylation(GO:0035574) |
| 0.0 |
0.1 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 |
0.2 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.2 |
GO:1990564 |
protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 |
0.1 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 |
0.3 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.0 |
GO:0021612 |
facial nerve structural organization(GO:0021612) |
| 0.0 |
0.6 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
1.5 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 |
0.1 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.0 |
0.1 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.2 |
GO:1904587 |
glycoprotein ERAD pathway(GO:0097466) response to glycoprotein(GO:1904587) |
| 0.0 |
0.1 |
GO:0046077 |
dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 |
0.1 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 |
0.2 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 |
0.6 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.1 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 |
0.2 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.9 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.0 |
0.1 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.0 |
0.3 |
GO:0034390 |
smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.0 |
0.2 |
GO:0009650 |
UV protection(GO:0009650) |
| 0.0 |
0.2 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0071500 |
cellular response to nitrosative stress(GO:0071500) |
| 0.0 |
0.1 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.0 |
0.1 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 |
0.7 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.3 |
GO:0060080 |
inhibitory postsynaptic potential(GO:0060080) |
| 0.0 |
0.2 |
GO:0014832 |
urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 |
0.1 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.0 |
0.2 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.3 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 |
0.1 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 |
0.4 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 |
0.1 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 |
0.1 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.0 |
0.1 |
GO:0032534 |
regulation of microvillus assembly(GO:0032534) |
| 0.0 |
0.1 |
GO:0031652 |
positive regulation of heat generation(GO:0031652) |
| 0.0 |
0.1 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.3 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.1 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.1 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.5 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.1 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 |
0.7 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.3 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.0 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.1 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 |
0.2 |
GO:0018126 |
protein hydroxylation(GO:0018126) |
| 0.0 |
0.2 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.0 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 |
0.1 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 |
0.2 |
GO:0021527 |
spinal cord association neuron differentiation(GO:0021527) |
| 0.0 |
0.1 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.0 |
0.4 |
GO:0007026 |
negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 |
0.1 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.5 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.4 |
GO:0048013 |
ephrin receptor signaling pathway(GO:0048013) |
| 0.0 |
0.3 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.0 |
0.5 |
GO:0007628 |
adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 |
0.0 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.0 |
0.1 |
GO:0014823 |
response to activity(GO:0014823) |
| 0.0 |
0.1 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |