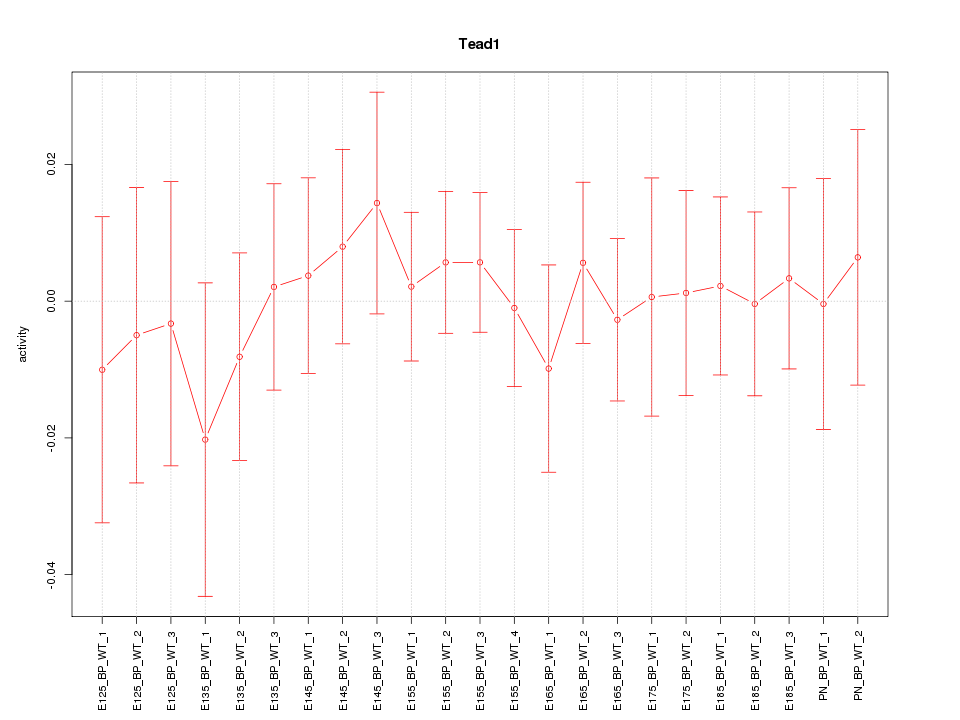
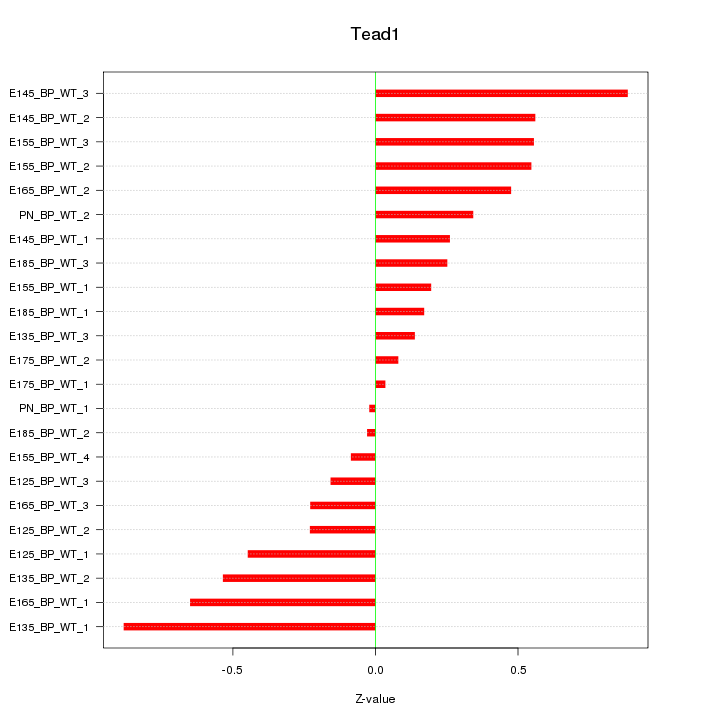
Motif ID: Tead1
Z-value: 0.422
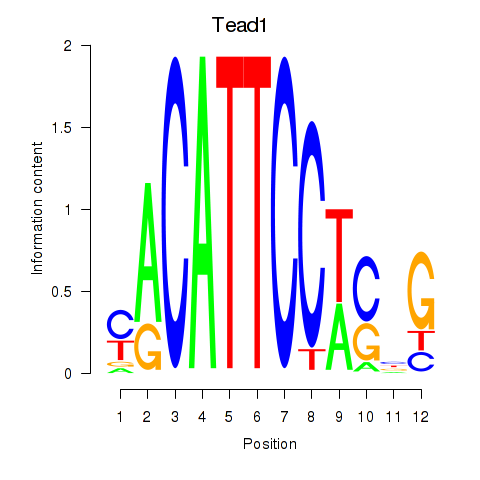
Transcription factors associated with Tead1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tead1 | ENSMUSG00000055320.10 | Tead1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tead1 | mm10_v2_chr7_+_112742025_112742060 | 0.29 | 1.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.2 | 1.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.7 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.1 | 0.4 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 1.0 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.8 | GO:1905206 | positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.1 | 0.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0009182 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 | 2.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.3 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 0.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.1 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.0 | 0.2 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 2.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) positive regulation of RNA binding(GO:1905216) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.0 | 0.8 | GO:0042058 | regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.3 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.9 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.7 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.5 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 2.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 1.3 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 1.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.7 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.1 | 0.7 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 1.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.9 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.0 | 0.1 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) alpha-1B adrenergic receptor binding(GO:0031692) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |