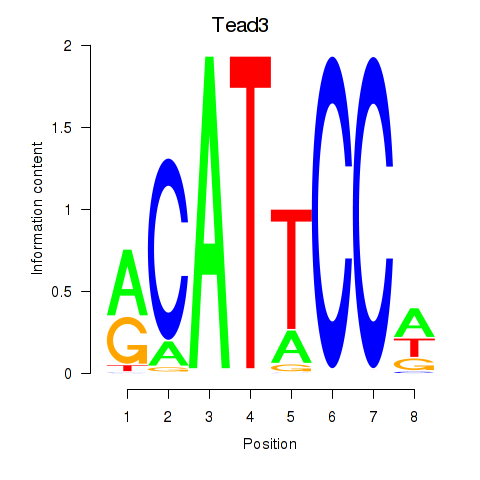
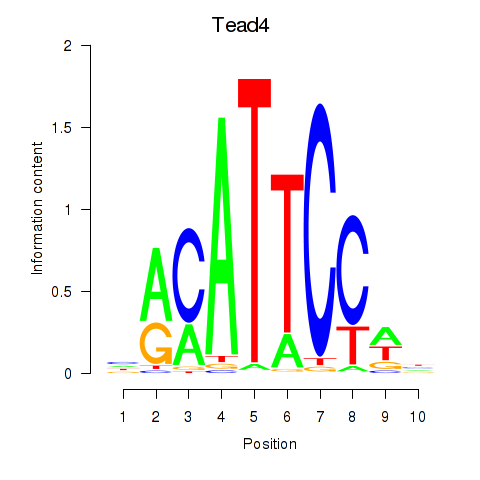
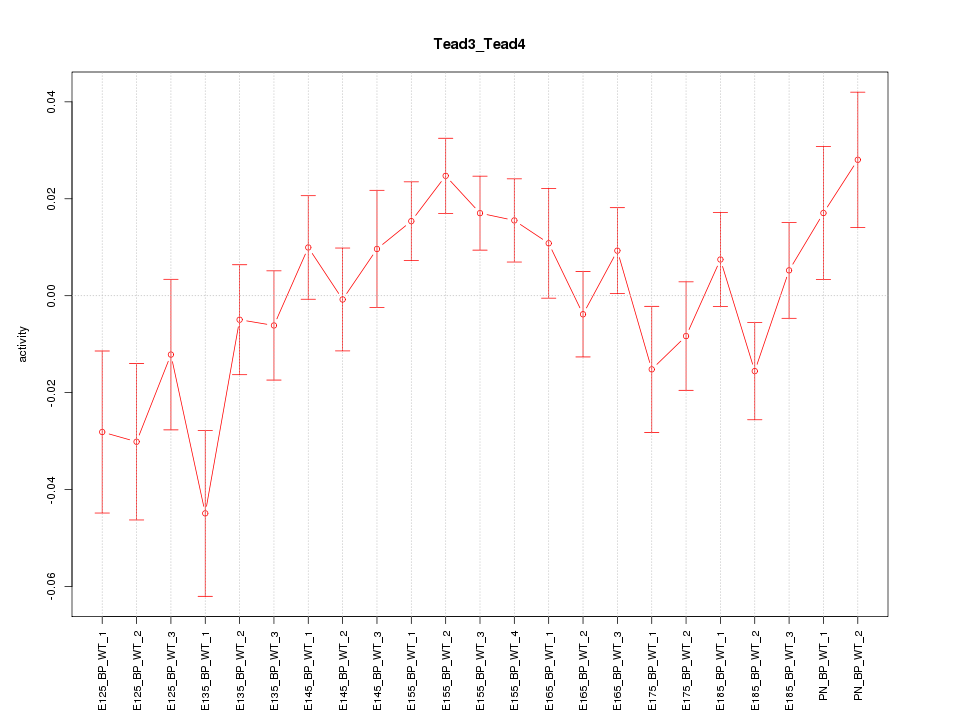
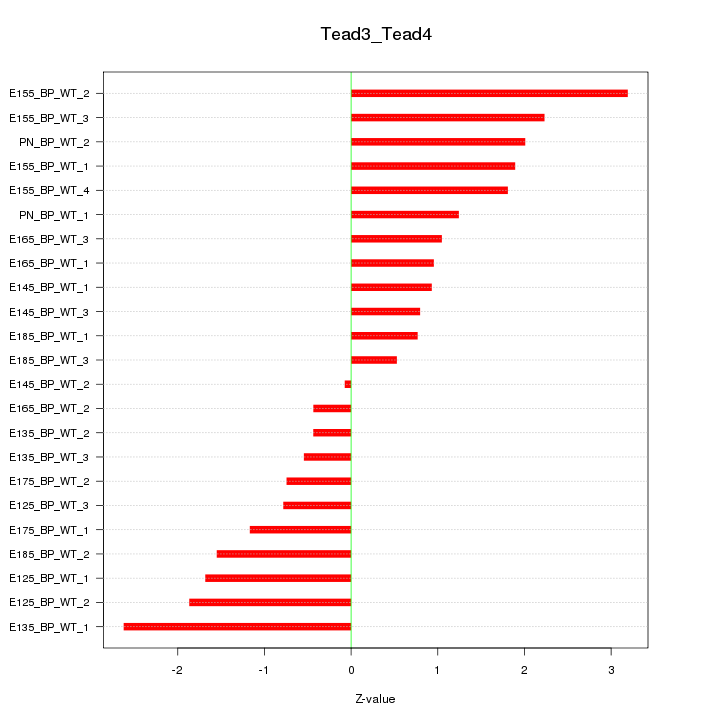
| 2.8 |
11.1 |
GO:0070318 |
positive regulation of G0 to G1 transition(GO:0070318) |
| 1.3 |
4.0 |
GO:2001139 |
negative regulation of postsynaptic membrane organization(GO:1901627) negative regulation of dendritic spine maintenance(GO:1902951) negative regulation of phospholipid efflux(GO:1902999) regulation of lipid transport across blood brain barrier(GO:1903000) negative regulation of lipid transport across blood brain barrier(GO:1903001) positive regulation of lipid transport across blood brain barrier(GO:1903002) negative regulation of phospholipid transport(GO:2001139) |
| 1.1 |
3.3 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.9 |
5.2 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.8 |
6.7 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.8 |
2.3 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.7 |
5.2 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.7 |
5.8 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.7 |
3.6 |
GO:2000298 |
regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.7 |
2.1 |
GO:1901228 |
positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.7 |
3.4 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.6 |
6.4 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.6 |
4.2 |
GO:1905206 |
positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.6 |
1.8 |
GO:0048866 |
stem cell fate specification(GO:0048866) |
| 0.5 |
1.6 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.5 |
2.7 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.5 |
1.6 |
GO:1902358 |
sulfate transmembrane transport(GO:1902358) |
| 0.5 |
3.1 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.5 |
1.4 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 0.5 |
2.3 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.4 |
2.7 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 |
3.0 |
GO:0072257 |
metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.4 |
1.2 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.4 |
1.6 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.4 |
1.5 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.4 |
1.5 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) |
| 0.4 |
1.1 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.4 |
1.5 |
GO:0042938 |
dipeptide transport(GO:0042938) |
| 0.4 |
1.1 |
GO:0061324 |
cardiac right atrium morphogenesis(GO:0003213) negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.3 |
0.3 |
GO:0061113 |
pancreas morphogenesis(GO:0061113) |
| 0.3 |
2.0 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 |
3.3 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.3 |
3.3 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
2.5 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.2 |
0.7 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 |
0.7 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.2 |
4.9 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.2 |
1.9 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.2 |
0.7 |
GO:0008065 |
establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.2 |
1.1 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 |
0.9 |
GO:0046487 |
glyoxylate metabolic process(GO:0046487) |
| 0.2 |
1.1 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.2 |
1.0 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.2 |
0.6 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.2 |
1.2 |
GO:0015879 |
carnitine transport(GO:0015879) |
| 0.2 |
0.4 |
GO:0072289 |
metanephric nephron tubule formation(GO:0072289) |
| 0.2 |
2.5 |
GO:1901629 |
regulation of presynaptic membrane organization(GO:1901629) |
| 0.2 |
0.5 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.2 |
1.2 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.2 |
1.2 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.2 |
0.5 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 |
0.7 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
1.8 |
GO:0031272 |
regulation of pseudopodium assembly(GO:0031272) |
| 0.1 |
0.4 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.1 |
0.6 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 |
0.7 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
1.8 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 |
0.7 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.4 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 |
0.4 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.1 |
1.2 |
GO:1902459 |
positive regulation of stem cell population maintenance(GO:1902459) |
| 0.1 |
1.9 |
GO:2000353 |
positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 |
0.6 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.7 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.8 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 |
0.6 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
1.5 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.7 |
GO:0017121 |
phospholipid scrambling(GO:0017121) |
| 0.1 |
0.6 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.1 |
0.6 |
GO:0060056 |
mammary gland involution(GO:0060056) |
| 0.1 |
0.4 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 |
1.3 |
GO:0030213 |
hyaluronan biosynthetic process(GO:0030213) |
| 0.1 |
0.2 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.2 |
GO:2000832 |
negative regulation of steroid hormone secretion(GO:2000832) negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 |
0.3 |
GO:1990414 |
replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 |
0.5 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 |
0.8 |
GO:0061469 |
regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 |
0.3 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 |
0.5 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 |
0.4 |
GO:2000210 |
positive regulation of anoikis(GO:2000210) |
| 0.1 |
0.5 |
GO:0010454 |
negative regulation of cell fate commitment(GO:0010454) |
| 0.1 |
0.7 |
GO:0060710 |
chorio-allantoic fusion(GO:0060710) |
| 0.1 |
0.6 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
8.2 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 |
0.2 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 |
1.5 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.1 |
4.8 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.1 |
0.9 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
0.7 |
GO:0003215 |
cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 |
0.3 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 |
0.4 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.2 |
GO:0090110 |
cargo loading into COPII-coated vesicle(GO:0090110) regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 |
0.7 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.0 |
0.9 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 |
0.4 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
2.3 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.6 |
GO:0014067 |
negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 |
1.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.1 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 |
1.4 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.3 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.0 |
1.0 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 |
2.1 |
GO:0042058 |
regulation of epidermal growth factor receptor signaling pathway(GO:0042058) |
| 0.0 |
1.3 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 |
1.5 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.4 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 |
0.5 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
1.0 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.3 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.3 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.4 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 |
1.3 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.5 |
GO:0070831 |
basement membrane assembly(GO:0070831) |
| 0.0 |
0.2 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.1 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 |
2.3 |
GO:0010811 |
positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 |
0.3 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 |
4.4 |
GO:0031032 |
actomyosin structure organization(GO:0031032) |
| 0.0 |
1.1 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 |
0.7 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.0 |
0.1 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 |
0.2 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 |
0.1 |
GO:1903847 |
regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 |
1.6 |
GO:0061077 |
chaperone-mediated protein folding(GO:0061077) |
| 0.0 |
0.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
1.5 |
GO:0030968 |
endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 |
0.1 |
GO:0070973 |
vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 |
0.1 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 |
0.8 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.2 |
GO:0002467 |
germinal center formation(GO:0002467) |
| 0.0 |
0.3 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.1 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.3 |
GO:0060438 |
trachea development(GO:0060438) |
| 0.0 |
1.7 |
GO:0000187 |
activation of MAPK activity(GO:0000187) |
| 0.0 |
1.4 |
GO:0019319 |
hexose biosynthetic process(GO:0019319) |
| 0.0 |
0.9 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.2 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
3.0 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.6 |
GO:0003407 |
neural retina development(GO:0003407) |
| 0.0 |
0.3 |
GO:0048635 |
negative regulation of muscle organ development(GO:0048635) |
| 0.0 |
0.7 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
1.1 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.3 |
GO:0044381 |
glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.0 |
0.2 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 |
1.4 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.0 |
0.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.0 |
0.1 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.3 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.0 |
0.6 |
GO:0060997 |
dendritic spine morphogenesis(GO:0060997) |