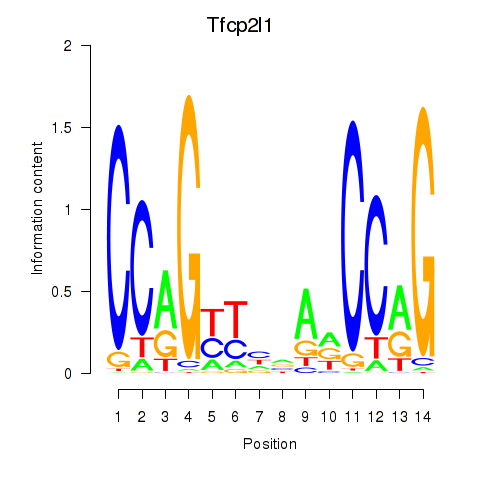
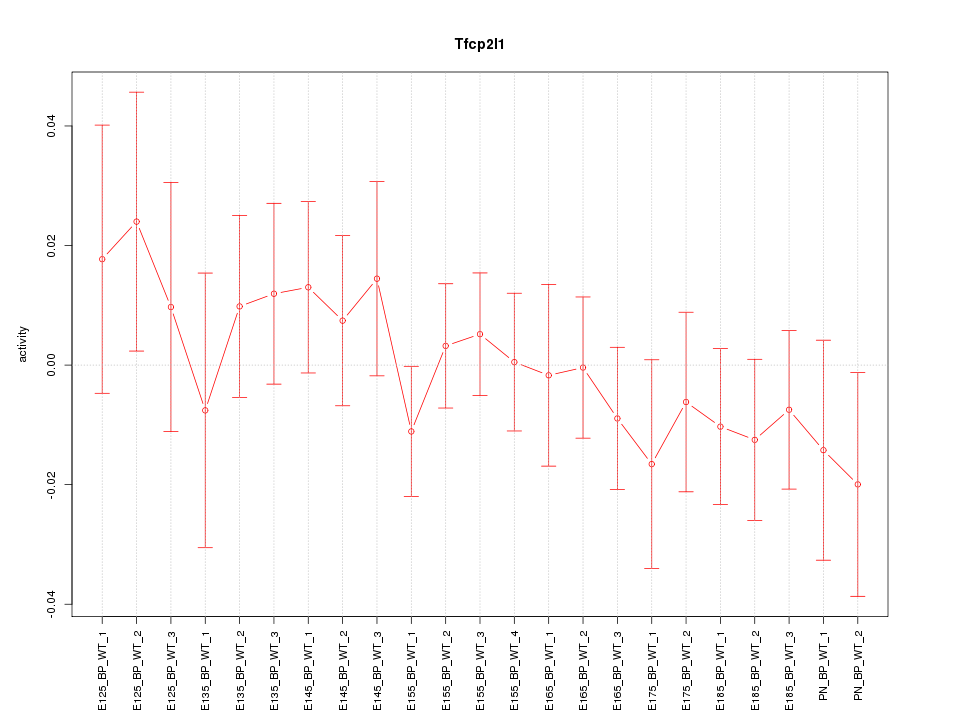
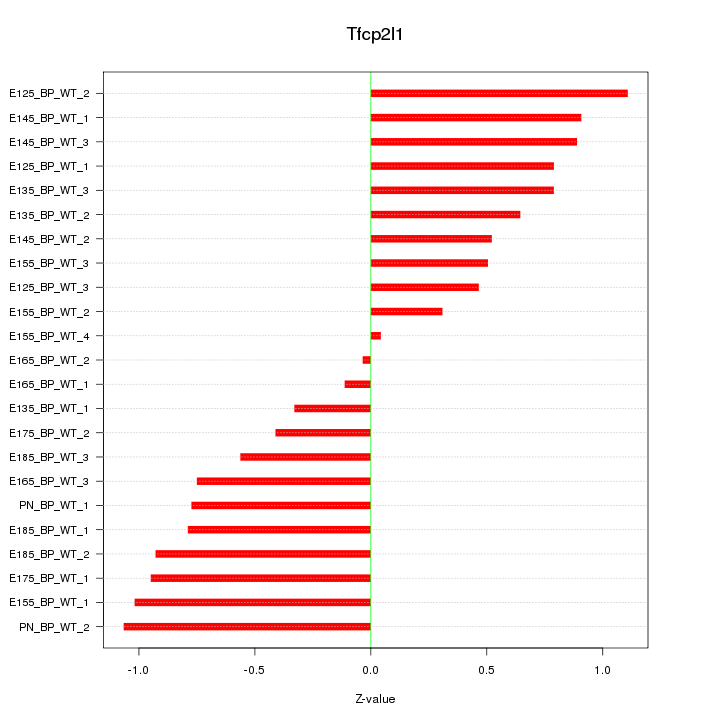
| 1.2 |
3.5 |
GO:0060598 |
dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.7 |
8.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.4 |
1.2 |
GO:0098528 |
terpenoid catabolic process(GO:0016115) skeletal muscle fiber differentiation(GO:0098528) |
| 0.4 |
1.1 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 0.3 |
0.8 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 |
0.7 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.2 |
0.5 |
GO:0002314 |
germinal center B cell differentiation(GO:0002314) positive regulation of germinal center formation(GO:0002636) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) purine nucleobase salvage(GO:0043096) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) xanthine metabolic process(GO:0046110) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.2 |
0.7 |
GO:0060266 |
negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 0.1 |
0.4 |
GO:2000195 |
negative regulation of female gonad development(GO:2000195) |
| 0.1 |
0.7 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.1 |
0.9 |
GO:0061436 |
establishment of skin barrier(GO:0061436) |
| 0.1 |
0.3 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 |
0.8 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.1 |
0.6 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 |
1.5 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.1 |
0.5 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.1 |
1.0 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
1.7 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.2 |
GO:1902915 |
negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 |
0.4 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.4 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
5.8 |
GO:0051028 |
mRNA transport(GO:0051028) |
| 0.0 |
0.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
1.6 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
2.3 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.1 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 0.0 |
0.5 |
GO:0043278 |
response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.0 |
0.2 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 |
0.4 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.0 |
1.3 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
1.3 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.4 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.1 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 |
0.5 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.6 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.9 |
GO:0030317 |
sperm motility(GO:0030317) |
| 0.0 |
0.7 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.3 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.1 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.1 |
GO:0070886 |
positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 |
0.5 |
GO:0090004 |
positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 |
0.2 |
GO:0071425 |
hematopoietic stem cell proliferation(GO:0071425) |