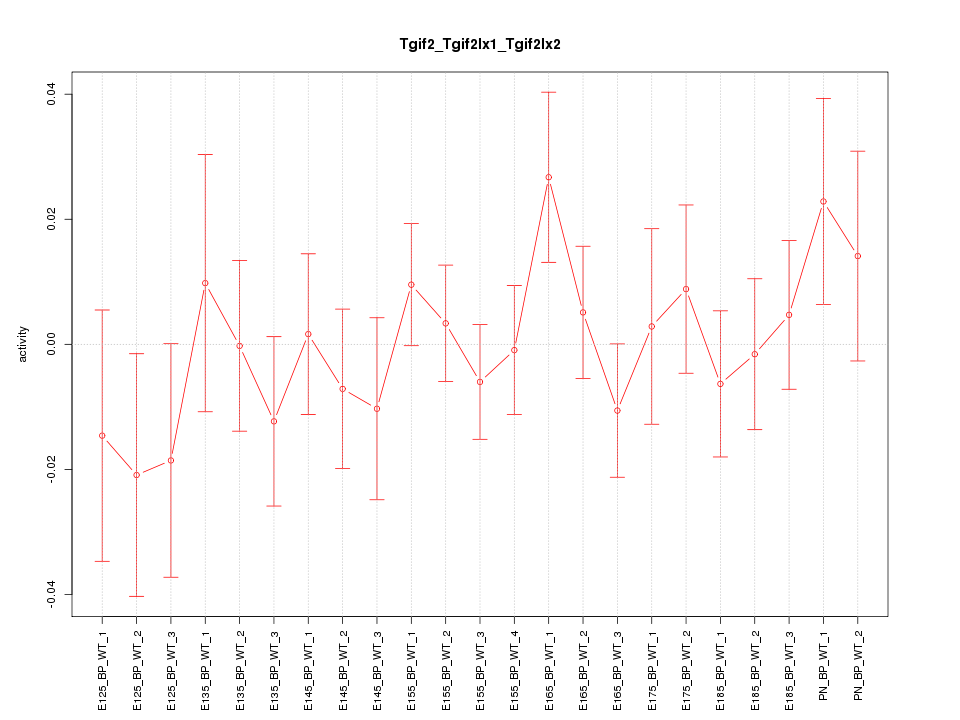
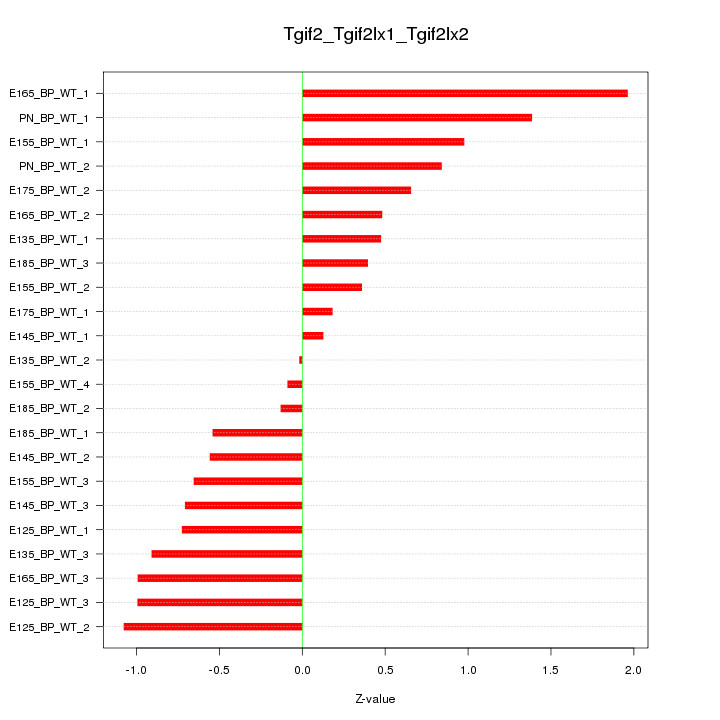
Motif ID: Tgif2_Tgif2lx1_Tgif2lx2
Z-value: 0.800
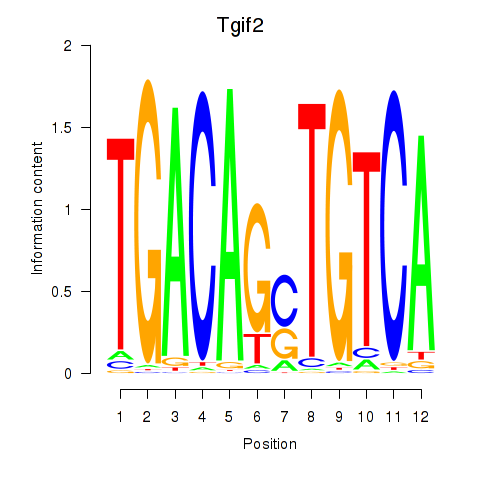
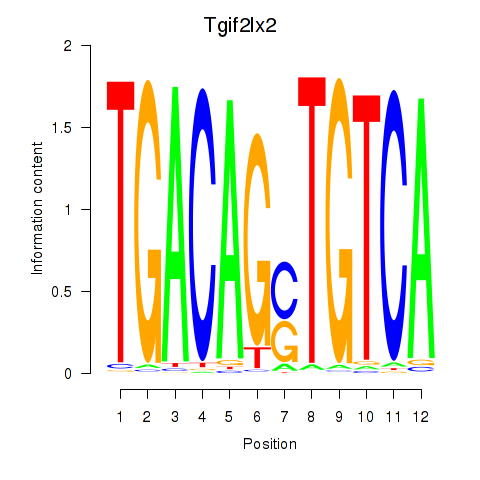
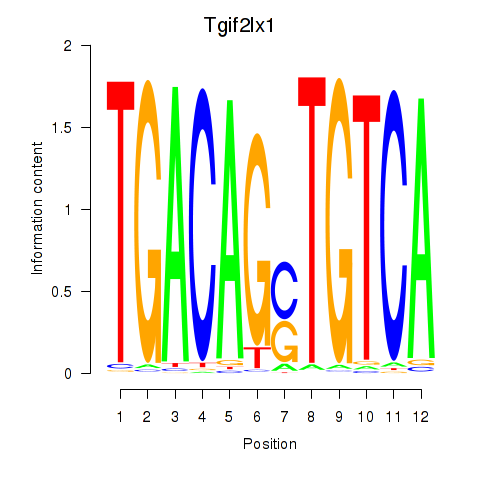
Transcription factors associated with Tgif2_Tgif2lx1_Tgif2lx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Tgif2 | ENSMUSG00000062175.7 | Tgif2 |
| Tgif2lx1 | ENSMUSG00000061283.4 | Tgif2lx1 |
| Tgif2lx2 | ENSMUSG00000063242.6 | Tgif2lx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tgif2 | mm10_v2_chr2_+_156840966_156841017 | 0.43 | 4.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.6 | 3.1 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 0.3 | 1.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.3 | 1.4 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 2.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.4 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.4 | GO:2001187 | positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.1 | 1.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.3 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.9 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 1.0 | GO:0007379 | segment specification(GO:0007379) |
| 0.1 | 5.5 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.3 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.4 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 1.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.6 | GO:0002591 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) |
| 0.1 | 0.2 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.1 | 1.2 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.1 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 1.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.6 | GO:0046459 | short-chain fatty acid metabolic process(GO:0046459) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.1 | 0.2 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 0.1 | 0.3 | GO:0042701 | progesterone secretion(GO:0042701) |
| 0.1 | 0.3 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.3 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.5 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 1.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.4 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.3 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 1.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.7 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.6 | GO:0001504 | neurotransmitter uptake(GO:0001504) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.8 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.0 | 0.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.7 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 1.8 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.0 | 0.4 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 1.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 1.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 1.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.9 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 13.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.3 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.4 | 1.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 3.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.2 | 2.9 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.2 | 1.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 1.4 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.7 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 1.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.6 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 1.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.3 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.1 | 0.4 | GO:0015125 | virus receptor activity(GO:0001618) bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.6 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.6 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 1.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.5 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 3.0 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |