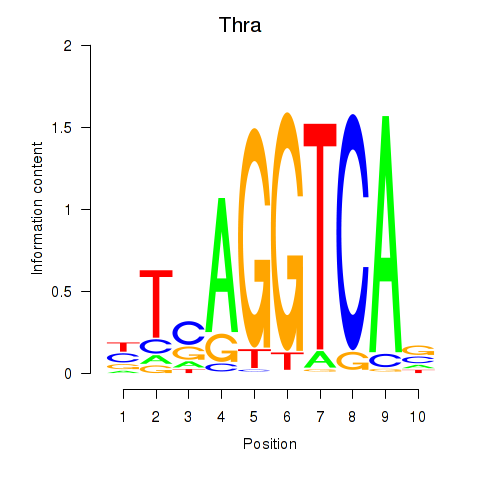
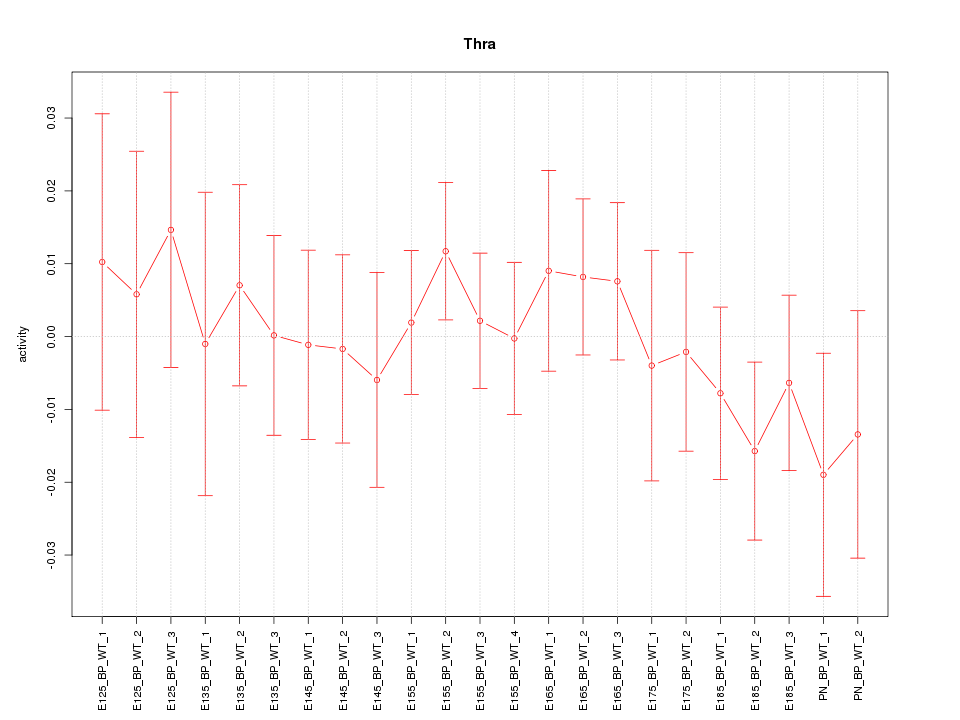
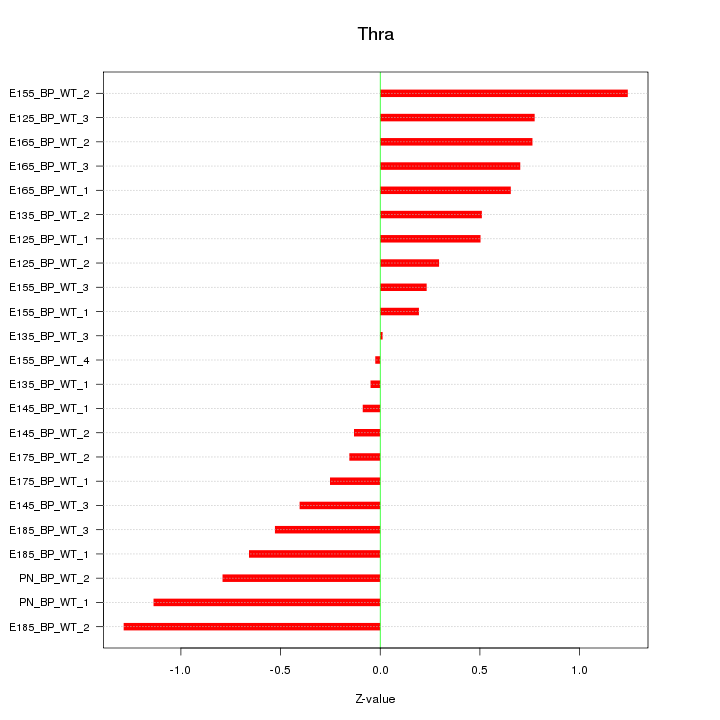
| 0.4 |
1.1 |
GO:0070093 |
negative regulation of glucagon secretion(GO:0070093) |
| 0.3 |
0.9 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 |
0.8 |
GO:0021849 |
neuroblast division in subventricular zone(GO:0021849) |
| 0.2 |
1.2 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.2 |
0.9 |
GO:0035426 |
extracellular matrix-cell signaling(GO:0035426) |
| 0.2 |
0.5 |
GO:0006657 |
CDP-choline pathway(GO:0006657) |
| 0.1 |
0.6 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 |
0.6 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.8 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.1 |
0.5 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
1.6 |
GO:0021979 |
hypothalamus cell differentiation(GO:0021979) |
| 0.1 |
0.7 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
0.4 |
GO:0055118 |
negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.1 |
0.7 |
GO:0035902 |
response to immobilization stress(GO:0035902) |
| 0.1 |
0.4 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) |
| 0.1 |
0.7 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 |
0.3 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.3 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.1 |
0.7 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.5 |
GO:0090038 |
negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 |
1.1 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.1 |
0.3 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 |
0.6 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 |
0.4 |
GO:1903753 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) hard palate development(GO:0060022) negative regulation of p38MAPK cascade(GO:1903753) |
| 0.1 |
0.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 |
1.0 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
0.3 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.1 |
0.2 |
GO:0090202 |
regulation of primitive erythrocyte differentiation(GO:0010725) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 |
0.3 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.0 |
0.2 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.0 |
0.3 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.2 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 |
0.2 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 |
0.2 |
GO:0071105 |
response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.0 |
0.2 |
GO:0051890 |
regulation of cardioblast differentiation(GO:0051890) |
| 0.0 |
0.1 |
GO:0007341 |
penetration of zona pellucida(GO:0007341) |
| 0.0 |
0.5 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.2 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.0 |
0.3 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.1 |
GO:0071929 |
alpha-tubulin acetylation(GO:0071929) |
| 0.0 |
0.3 |
GO:0019240 |
citrulline biosynthetic process(GO:0019240) |
| 0.0 |
0.5 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.1 |
GO:0046341 |
CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 |
0.4 |
GO:0032463 |
negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 |
0.1 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 0.0 |
0.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.1 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 |
0.2 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.2 |
GO:0051004 |
plasma membrane to endosome transport(GO:0048227) regulation of lipoprotein lipase activity(GO:0051004) |
| 0.0 |
0.1 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.0 |
0.4 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.6 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.0 |
0.2 |
GO:0060484 |
lung-associated mesenchyme development(GO:0060484) |
| 0.0 |
0.4 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.0 |
0.2 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.0 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.0 |
0.5 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.5 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.0 |
0.7 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.0 |
0.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.2 |
GO:0090435 |
protein localization to nuclear envelope(GO:0090435) |
| 0.0 |
0.1 |
GO:0061084 |
negative regulation of protein refolding(GO:0061084) |
| 0.0 |
0.2 |
GO:0035457 |
cellular response to interferon-alpha(GO:0035457) type I interferon biosynthetic process(GO:0045351) positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 |
0.3 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.1 |
GO:0042473 |
production of siRNA involved in RNA interference(GO:0030422) outer ear morphogenesis(GO:0042473) |
| 0.0 |
0.4 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.2 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 |
0.2 |
GO:0002385 |
mucosal immune response(GO:0002385) |
| 0.0 |
0.0 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.0 |
0.1 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.6 |
GO:0043392 |
negative regulation of DNA binding(GO:0043392) |
| 0.0 |
0.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 |
0.2 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |