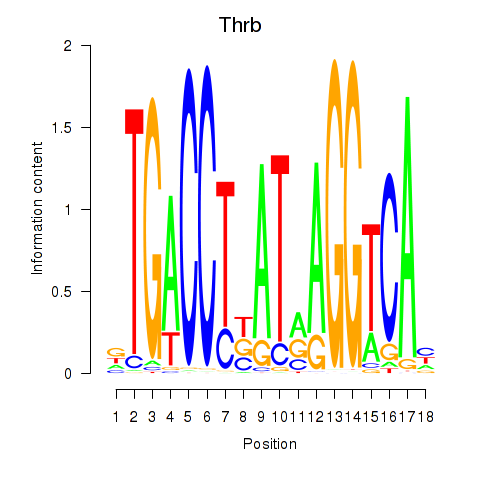
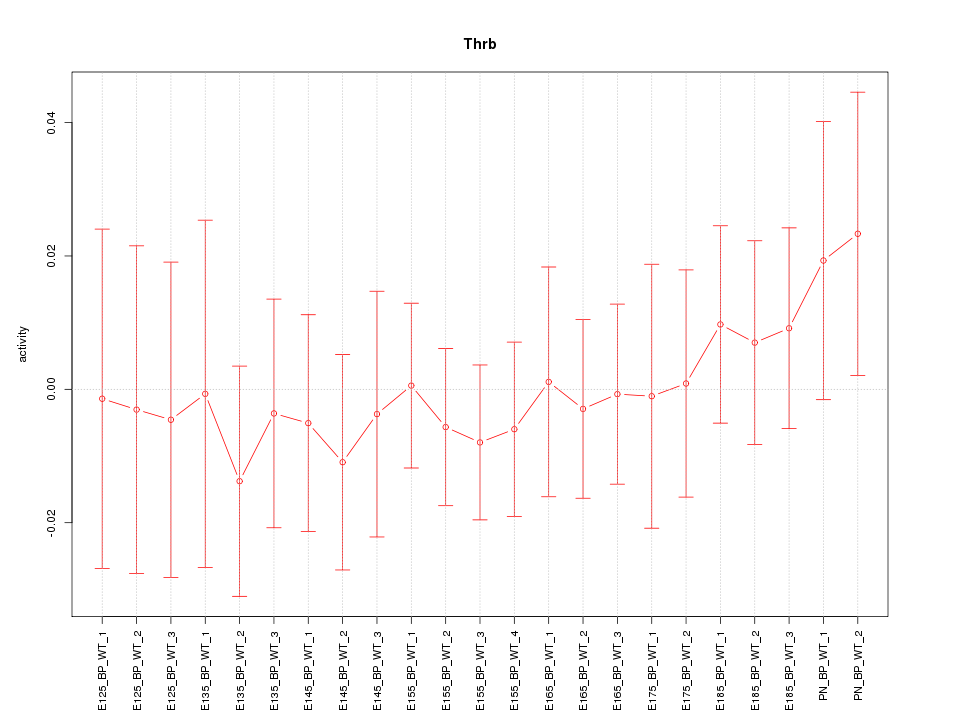
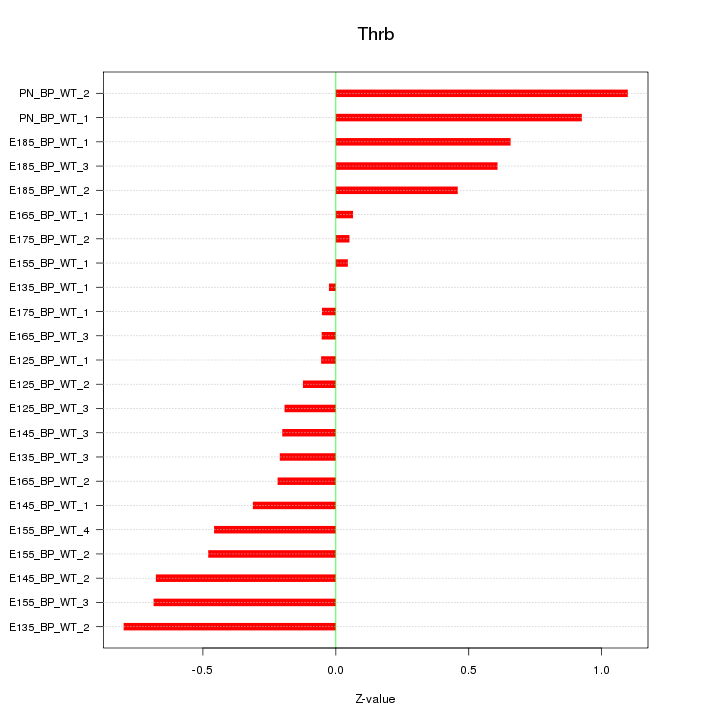
| 1.3 |
5.3 |
GO:0004667 |
prostaglandin-D synthase activity(GO:0004667) |
| 0.3 |
1.4 |
GO:0015186 |
L-asparagine transmembrane transporter activity(GO:0015182) L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 |
0.4 |
GO:0048045 |
4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.1 |
1.2 |
GO:0016151 |
nickel cation binding(GO:0016151) |
| 0.1 |
0.7 |
GO:0018741 |
alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 |
0.3 |
GO:0004965 |
G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 |
0.7 |
GO:0005388 |
calcium-transporting ATPase activity(GO:0005388) |
| 0.0 |
0.7 |
GO:0043747 |
protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.0 |
0.8 |
GO:0016709 |
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 |
0.5 |
GO:0008242 |
omega peptidase activity(GO:0008242) |
| 0.0 |
0.2 |
GO:0016863 |
intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 |
0.1 |
GO:0032405 |
MutLalpha complex binding(GO:0032405) MutSalpha complex binding(GO:0032407) |
| 0.0 |
1.9 |
GO:0051015 |
actin filament binding(GO:0051015) |
| 0.0 |
0.2 |
GO:0005351 |
sugar:proton symporter activity(GO:0005351) |