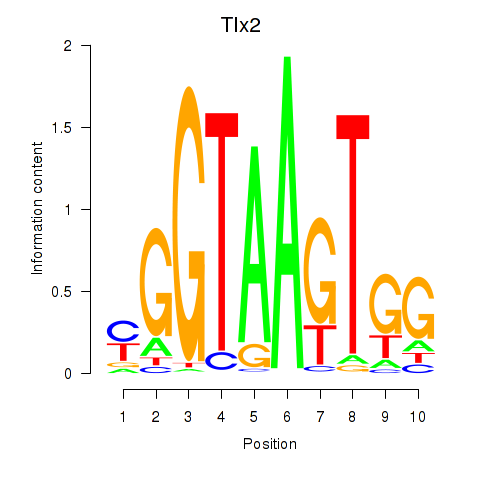
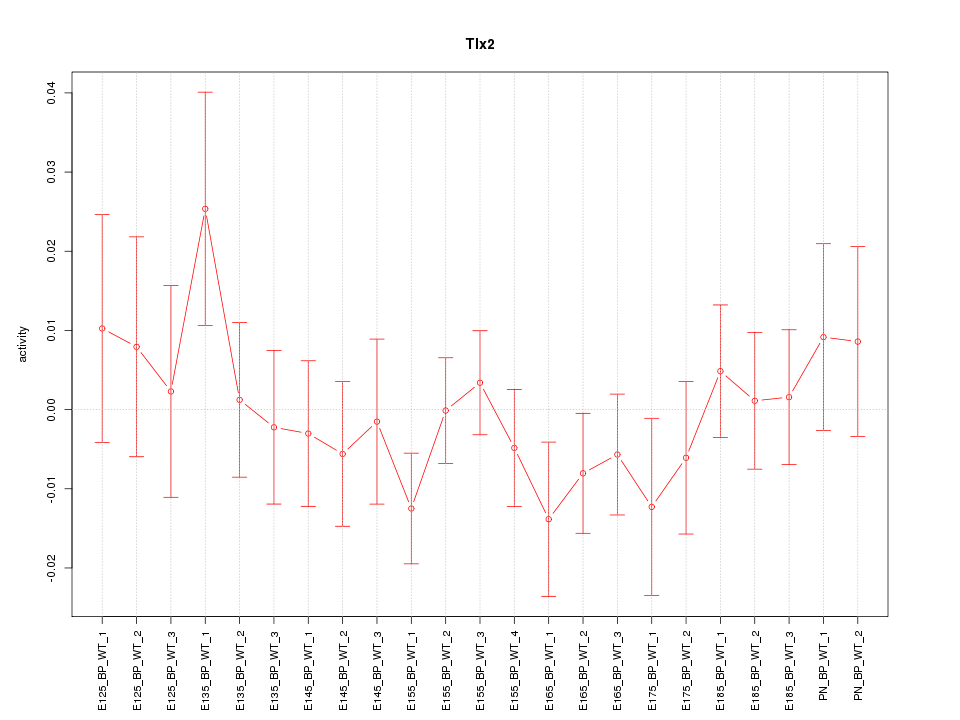
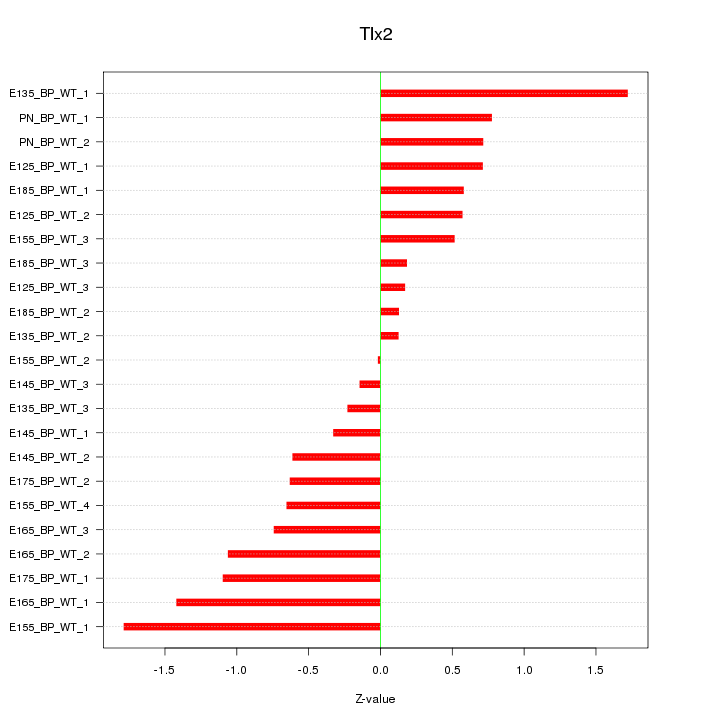
| 0.8 |
7.0 |
GO:1902715 |
positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.8 |
2.3 |
GO:1903048 |
regulation of acetylcholine-gated cation channel activity(GO:1903048) |
| 0.7 |
2.7 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.6 |
1.8 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.5 |
1.6 |
GO:2000562 |
negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.4 |
0.9 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.3 |
0.7 |
GO:0060455 |
negative regulation of gastric acid secretion(GO:0060455) |
| 0.3 |
1.3 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 |
0.9 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.3 |
0.8 |
GO:0035519 |
protein K29-linked ubiquitination(GO:0035519) |
| 0.3 |
1.3 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.3 |
0.8 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 |
0.7 |
GO:0090187 |
zymogen granule exocytosis(GO:0070625) positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.2 |
0.7 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
0.7 |
GO:0018298 |
protein-chromophore linkage(GO:0018298) |
| 0.2 |
1.3 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.2 |
0.5 |
GO:0032513 |
negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.2 |
0.5 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 |
0.5 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.2 |
0.6 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 |
0.5 |
GO:0010752 |
signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 0.2 |
0.5 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.2 |
0.5 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 0.2 |
1.6 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.2 |
1.1 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.9 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.1 |
0.9 |
GO:0002934 |
desmosome organization(GO:0002934) |
| 0.1 |
0.1 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.1 |
0.8 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 |
0.4 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.1 |
0.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.5 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.1 |
0.9 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.1 |
0.3 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 |
0.9 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.1 |
0.4 |
GO:0097709 |
connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 |
0.4 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 |
0.6 |
GO:0097491 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 |
0.4 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 |
0.2 |
GO:0002322 |
B cell proliferation involved in immune response(GO:0002322) |
| 0.1 |
0.9 |
GO:0032306 |
regulation of prostaglandin secretion(GO:0032306) |
| 0.1 |
0.5 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.1 |
0.7 |
GO:0000022 |
mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 |
0.5 |
GO:0043652 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) engulfment of apoptotic cell(GO:0043652) |
| 0.1 |
1.0 |
GO:0034312 |
diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.1 |
0.4 |
GO:0043973 |
histone H3-K4 acetylation(GO:0043973) |
| 0.1 |
0.5 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.1 |
0.4 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 |
0.4 |
GO:2000074 |
regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 |
0.2 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.1 |
0.4 |
GO:0030242 |
pexophagy(GO:0030242) |
| 0.1 |
0.2 |
GO:0046110 |
xanthine metabolic process(GO:0046110) |
| 0.1 |
0.3 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 |
0.3 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 |
0.2 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
0.9 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
0.2 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 |
0.3 |
GO:0045918 |
negative regulation of cytolysis(GO:0045918) |
| 0.1 |
0.7 |
GO:0032525 |
somite rostral/caudal axis specification(GO:0032525) |
| 0.1 |
0.4 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.1 |
0.2 |
GO:0045039 |
protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 |
0.2 |
GO:0070488 |
neutrophil aggregation(GO:0070488) |
| 0.1 |
0.9 |
GO:0014049 |
positive regulation of glutamate secretion(GO:0014049) |
| 0.1 |
0.3 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 |
0.2 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.1 |
0.5 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.5 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 |
0.5 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 |
0.2 |
GO:0007521 |
muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.0 |
0.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.4 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 |
0.0 |
GO:0071374 |
cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 |
0.7 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.2 |
GO:0018171 |
peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) |
| 0.0 |
0.6 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 |
0.9 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.0 |
0.4 |
GO:0042435 |
indole-containing compound biosynthetic process(GO:0042435) |
| 0.0 |
0.3 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.3 |
GO:0007343 |
egg activation(GO:0007343) |
| 0.0 |
0.9 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 0.0 |
0.1 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 |
1.0 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.2 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.5 |
GO:0007100 |
mitotic centrosome separation(GO:0007100) |
| 0.0 |
0.4 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.1 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.0 |
0.8 |
GO:0033622 |
integrin activation(GO:0033622) |
| 0.0 |
0.7 |
GO:0014046 |
dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 |
0.1 |
GO:0010593 |
negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 |
0.4 |
GO:0006122 |
mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 |
0.8 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.2 |
GO:0090400 |
stress-induced premature senescence(GO:0090400) |
| 0.0 |
0.3 |
GO:0002862 |
negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 |
0.2 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.4 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.0 |
0.1 |
GO:0071639 |
positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 |
0.6 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.1 |
GO:0070844 |
misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 |
0.4 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.4 |
GO:1904948 |
midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.0 |
1.0 |
GO:0007099 |
centriole replication(GO:0007099) |
| 0.0 |
0.4 |
GO:1903963 |
arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 |
0.1 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.0 |
0.1 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 |
0.6 |
GO:0014829 |
vascular smooth muscle contraction(GO:0014829) |
| 0.0 |
0.3 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 0.0 |
0.2 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.0 |
0.5 |
GO:0031110 |
regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 |
0.2 |
GO:0060343 |
trabecula formation(GO:0060343) |
| 0.0 |
0.1 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 |
0.2 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 |
0.3 |
GO:0007220 |
Notch receptor processing(GO:0007220) |
| 0.0 |
0.1 |
GO:2001181 |
positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.7 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
0.0 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.0 |
0.1 |
GO:0061732 |
acetyl-CoA biosynthetic process from pyruvate(GO:0006086) mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 |
0.0 |
GO:0034729 |
histone H3-K79 methylation(GO:0034729) |
| 0.0 |
0.3 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 |
0.2 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.5 |
GO:1904355 |
positive regulation of telomere capping(GO:1904355) |
| 0.0 |
1.2 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.1 |
GO:0044565 |
dendritic cell proliferation(GO:0044565) |
| 0.0 |
0.2 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.0 |
0.1 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 |
0.2 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
1.0 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.0 |
0.1 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.2 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.2 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 |
0.1 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 0.0 |
0.1 |
GO:0007320 |
insemination(GO:0007320) |
| 0.0 |
0.1 |
GO:2000383 |
regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 |
0.4 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.0 |
0.2 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.4 |
GO:0016082 |
synaptic vesicle priming(GO:0016082) |
| 0.0 |
0.4 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.0 |
GO:0002408 |
myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.0 |
1.5 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.2 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.6 |
GO:0060291 |
long-term synaptic potentiation(GO:0060291) |
| 0.0 |
0.2 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.2 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.6 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.2 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.1 |
GO:0030174 |
regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 |
0.3 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.1 |
GO:2000015 |
regulation of determination of dorsal identity(GO:2000015) |
| 0.0 |
0.5 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.2 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.0 |
0.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.2 |
GO:0045026 |
plasma membrane fusion(GO:0045026) |
| 0.0 |
0.4 |
GO:0035455 |
response to interferon-alpha(GO:0035455) |
| 0.0 |
0.2 |
GO:0043030 |
regulation of macrophage activation(GO:0043030) |
| 0.0 |
0.4 |
GO:0018105 |
peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 |
0.1 |
GO:0019400 |
glycerol metabolic process(GO:0006071) alditol metabolic process(GO:0019400) |
| 0.0 |
0.3 |
GO:0046173 |
polyol biosynthetic process(GO:0046173) |
| 0.0 |
0.4 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.4 |
GO:0034260 |
negative regulation of GTPase activity(GO:0034260) |
| 0.0 |
0.3 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 |
0.1 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 |
0.4 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.1 |
GO:1900364 |
negative regulation of mRNA 3'-end processing(GO:0031441) regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 |
0.3 |
GO:0006907 |
pinocytosis(GO:0006907) |
| 0.0 |
0.4 |
GO:0033119 |
negative regulation of RNA splicing(GO:0033119) |
| 0.0 |
0.3 |
GO:0043153 |
photoperiodism(GO:0009648) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.3 |
GO:0006346 |
methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 |
0.1 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.0 |
0.2 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.0 |
0.3 |
GO:0000413 |
protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 |
0.2 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.0 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 |
0.3 |
GO:0046847 |
filopodium assembly(GO:0046847) |
| 0.0 |
0.5 |
GO:0046777 |
protein autophosphorylation(GO:0046777) |
| 0.0 |
0.6 |
GO:0007093 |
mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 |
0.5 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
1.2 |
GO:0016052 |
carbohydrate catabolic process(GO:0016052) |
| 0.0 |
0.1 |
GO:0033262 |
regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.0 |
1.6 |
GO:0000082 |
G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.0 |
0.2 |
GO:0050832 |
defense response to fungus(GO:0050832) |
| 0.0 |
1.1 |
GO:0000086 |
G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 |
0.1 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.4 |
GO:0007566 |
embryo implantation(GO:0007566) |
| 0.0 |
0.2 |
GO:0051497 |
negative regulation of stress fiber assembly(GO:0051497) |