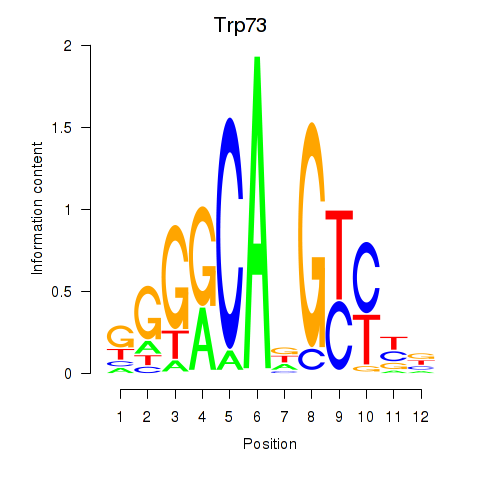
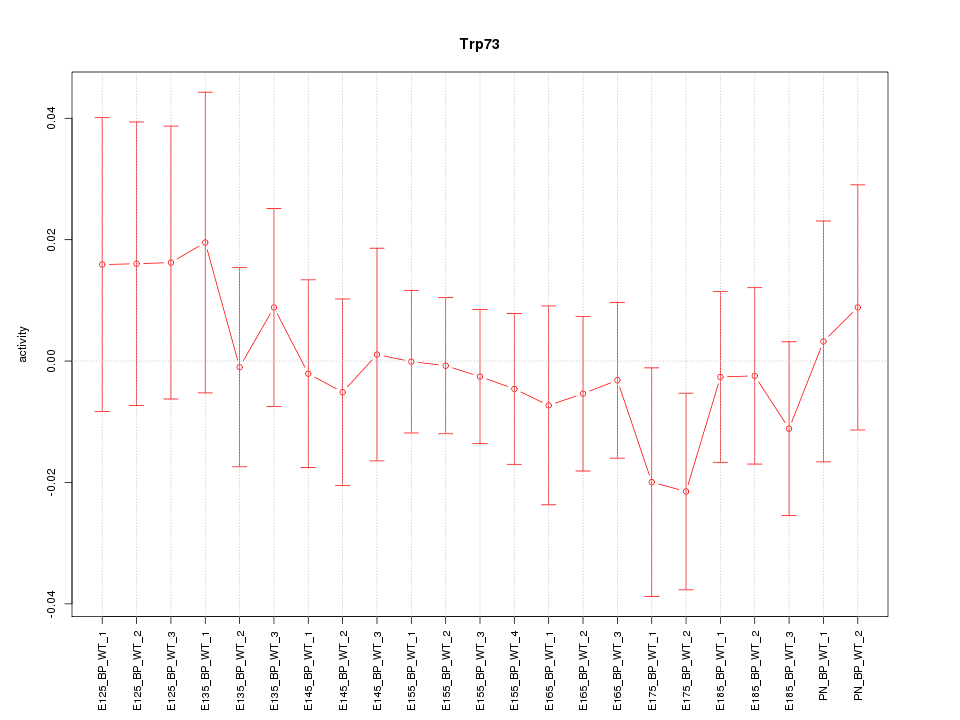
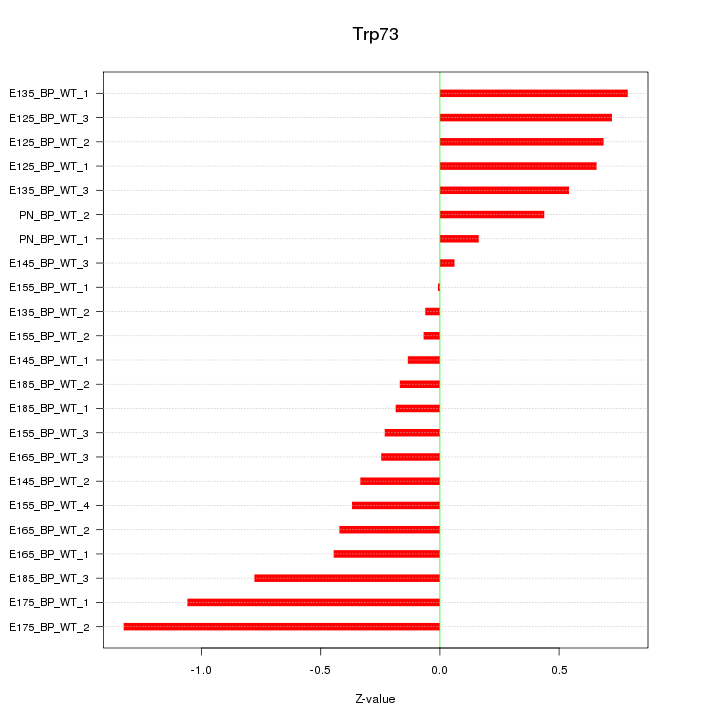
| 0.5 |
2.0 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.4 |
1.7 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 |
2.0 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.2 |
0.7 |
GO:0033566 |
gamma-tubulin complex localization(GO:0033566) |
| 0.2 |
1.8 |
GO:0071493 |
cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.2 |
1.0 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.2 |
0.6 |
GO:0032877 |
positive regulation of DNA endoreduplication(GO:0032877) |
| 0.2 |
2.2 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.2 |
2.1 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 |
0.4 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 |
0.6 |
GO:0060316 |
positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 |
0.4 |
GO:0043456 |
regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 |
0.6 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.3 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.1 |
1.1 |
GO:0031017 |
exocrine pancreas development(GO:0031017) |
| 0.1 |
1.5 |
GO:0042104 |
positive regulation of activated T cell proliferation(GO:0042104) |
| 0.1 |
0.2 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 |
0.2 |
GO:1903795 |
regulation of inorganic anion transmembrane transport(GO:1903795) |
| 0.0 |
0.5 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 |
0.3 |
GO:1900194 |
negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 |
0.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.2 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.0 |
1.2 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.2 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.5 |
GO:0045948 |
positive regulation of translational initiation(GO:0045948) |
| 0.0 |
0.4 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.2 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.1 |
GO:0090365 |
regulation of mRNA modification(GO:0090365) |
| 0.0 |
0.2 |
GO:0042523 |
positive regulation of tyrosine phosphorylation of Stat5 protein(GO:0042523) |
| 0.0 |
0.3 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 |
0.1 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.0 |
0.2 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
1.0 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.0 |
0.1 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 |
0.5 |
GO:0018279 |
protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 |
0.3 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |