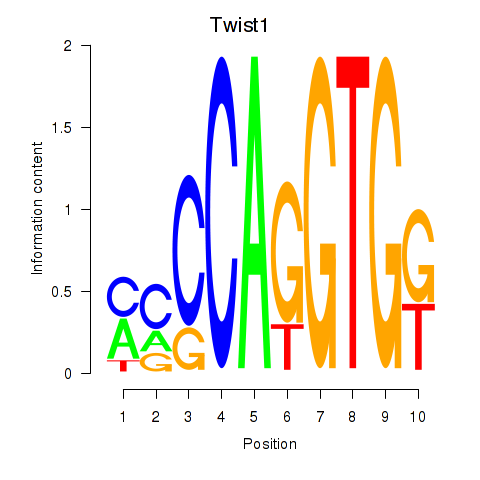
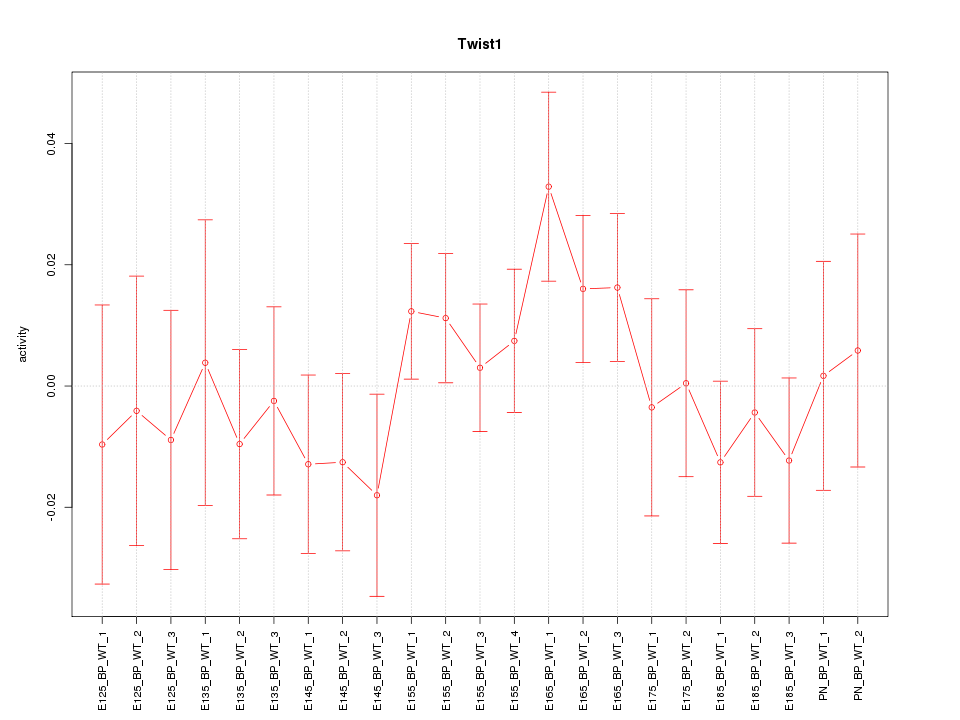
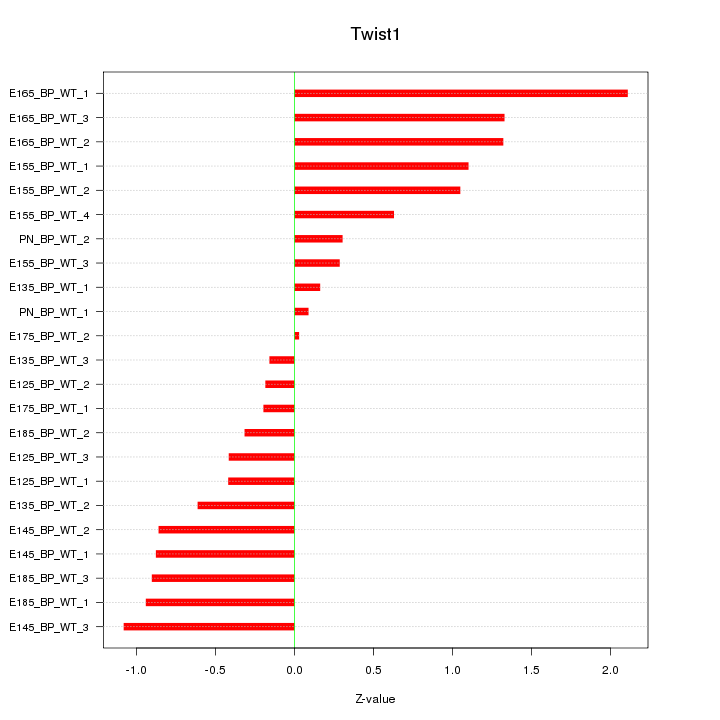
| 0.7 |
2.2 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.6 |
2.5 |
GO:0060032 |
notochord regression(GO:0060032) |
| 0.3 |
0.3 |
GO:1904173 |
regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.2 |
0.7 |
GO:0061324 |
mammary placode formation(GO:0060596) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.2 |
0.7 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 |
0.6 |
GO:1904395 |
regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.2 |
0.6 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 |
0.5 |
GO:0033860 |
regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.1 |
0.4 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
0.7 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 |
0.7 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 |
0.8 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
0.3 |
GO:0070103 |
interleukin-4-mediated signaling pathway(GO:0035771) tyrosine phosphorylation of Stat6 protein(GO:0042505) regulation of tyrosine phosphorylation of Stat6 protein(GO:0042525) regulation of interleukin-6-mediated signaling pathway(GO:0070103) negative regulation of interleukin-6-mediated signaling pathway(GO:0070104) |
| 0.1 |
0.4 |
GO:0072697 |
protein localization to cell cortex(GO:0072697) |
| 0.1 |
0.5 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
1.5 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.1 |
0.2 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.1 |
0.7 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
2.1 |
GO:0060445 |
branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 |
0.8 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
0.4 |
GO:0051503 |
adenine nucleotide transport(GO:0051503) |
| 0.1 |
0.5 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.2 |
GO:0051311 |
spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.0 |
0.2 |
GO:0060075 |
regulation of resting membrane potential(GO:0060075) |
| 0.0 |
0.1 |
GO:0003100 |
regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.0 |
0.2 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.4 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.2 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 |
0.3 |
GO:0070445 |
radial glial cell differentiation(GO:0060019) oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.4 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.1 |
GO:1904469 |
MyD88-independent toll-like receptor signaling pathway(GO:0002756) positive regulation of tumor necrosis factor secretion(GO:1904469) |
| 0.0 |
0.3 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 |
0.1 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 |
0.6 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.3 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.5 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.9 |
GO:0000038 |
very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 |
0.3 |
GO:0060766 |
negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 |
0.7 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) neutral amino acid transport(GO:0015804) |
| 0.0 |
1.1 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.7 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.4 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 |
0.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
0.2 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.0 |
0.2 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.0 |
0.3 |
GO:0030488 |
tRNA methylation(GO:0030488) |
| 0.0 |
0.1 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.5 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.0 |
0.3 |
GO:0040015 |
negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 |
0.1 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 |
0.3 |
GO:0048515 |
spermatid differentiation(GO:0048515) |