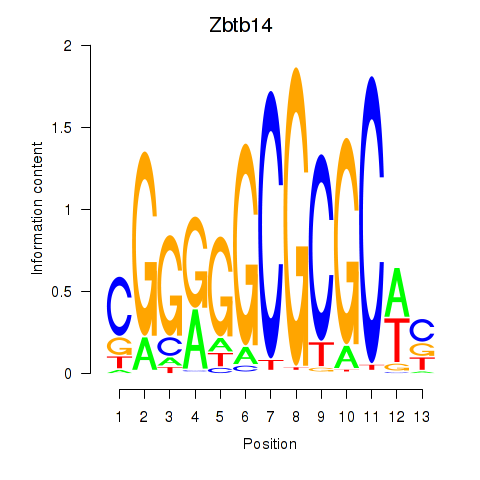
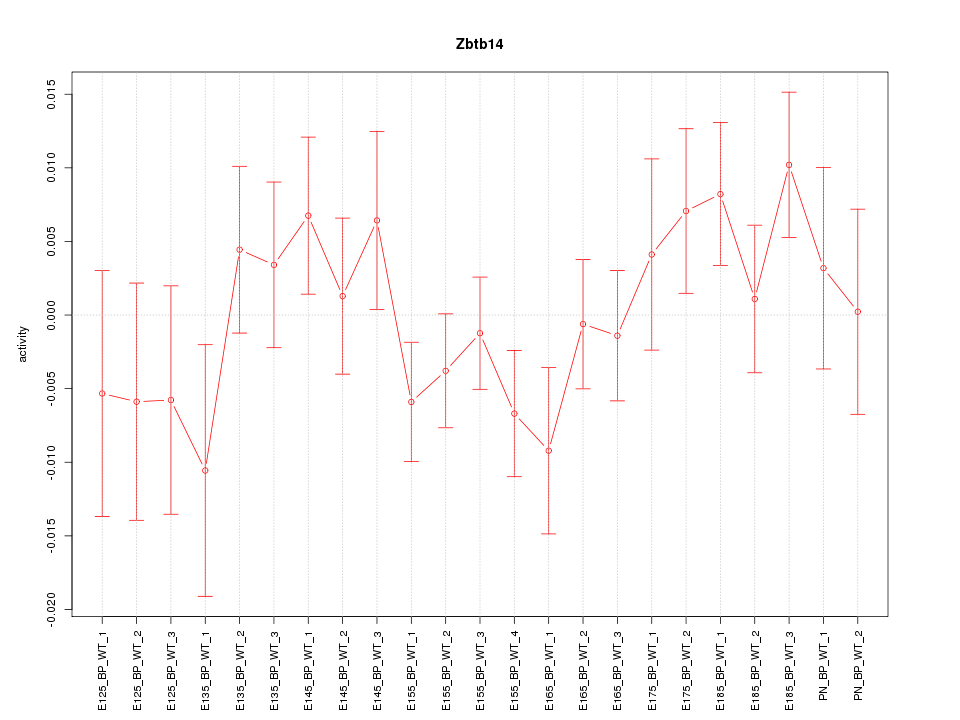
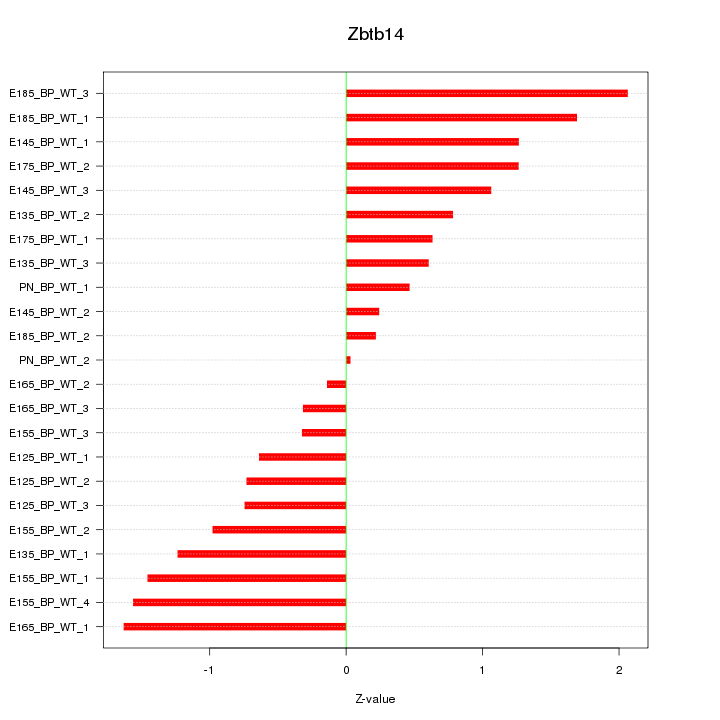
| 0.8 |
2.5 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.5 |
1.6 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.5 |
0.5 |
GO:0033024 |
mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.5 |
1.5 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 |
2.8 |
GO:0090336 |
positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.4 |
2.4 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.4 |
6.0 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.4 |
1.1 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.4 |
1.5 |
GO:0032788 |
saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.4 |
1.5 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.3 |
1.3 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.3 |
1.0 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.3 |
2.2 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 |
0.9 |
GO:1900673 |
olefin metabolic process(GO:1900673) |
| 0.3 |
0.9 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.3 |
1.7 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.3 |
0.8 |
GO:0043988 |
histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 |
0.3 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 0.3 |
1.1 |
GO:0044268 |
multicellular organismal protein metabolic process(GO:0044268) |
| 0.2 |
1.0 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 |
1.7 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 |
0.7 |
GO:1900149 |
positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 |
0.2 |
GO:0006931 |
substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 |
0.6 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 0.2 |
1.1 |
GO:0006447 |
regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 |
0.6 |
GO:0009193 |
pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.2 |
1.0 |
GO:0045657 |
positive regulation of monocyte differentiation(GO:0045657) |
| 0.2 |
0.6 |
GO:0006507 |
GPI anchor release(GO:0006507) |
| 0.2 |
1.4 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.2 |
0.5 |
GO:0097500 |
receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.2 |
0.8 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.2 |
1.2 |
GO:0061042 |
vascular wound healing(GO:0061042) |
| 0.2 |
0.5 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.2 |
1.0 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.2 |
0.5 |
GO:0010693 |
negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 |
0.6 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.2 |
2.0 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.2 |
1.7 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
0.4 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.1 |
0.1 |
GO:0038028 |
insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 |
0.7 |
GO:0032237 |
activation of store-operated calcium channel activity(GO:0032237) |
| 0.1 |
0.6 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) |
| 0.1 |
1.3 |
GO:0043615 |
astrocyte cell migration(GO:0043615) |
| 0.1 |
1.9 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
0.4 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 |
0.1 |
GO:0001922 |
B-1 B cell homeostasis(GO:0001922) |
| 0.1 |
0.3 |
GO:0050703 |
interleukin-1 alpha secretion(GO:0050703) |
| 0.1 |
0.4 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 |
0.1 |
GO:0070366 |
regulation of hepatocyte differentiation(GO:0070366) |
| 0.1 |
1.6 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 |
1.9 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.4 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.2 |
GO:0090427 |
activation of meiosis(GO:0090427) |
| 0.1 |
0.4 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.1 |
0.3 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 |
0.3 |
GO:0030210 |
heparin biosynthetic process(GO:0030210) |
| 0.1 |
0.3 |
GO:2001013 |
epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.1 |
0.8 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.1 |
0.3 |
GO:0051892 |
negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 |
2.2 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 |
1.1 |
GO:0014010 |
Schwann cell proliferation(GO:0014010) |
| 0.1 |
0.3 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.1 |
0.3 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.1 |
1.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 |
0.6 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
3.2 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 |
0.5 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 |
0.3 |
GO:0060214 |
stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) endocardium formation(GO:0060214) |
| 0.1 |
0.3 |
GO:1900041 |
intestinal D-glucose absorption(GO:0001951) negative regulation of interleukin-2 secretion(GO:1900041) terminal web assembly(GO:1902896) |
| 0.1 |
0.3 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 0.1 |
0.8 |
GO:0090219 |
negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 |
0.5 |
GO:0046598 |
positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 |
0.4 |
GO:0071692 |
protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) |
| 0.1 |
0.2 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.1 |
0.3 |
GO:0021546 |
rhombomere development(GO:0021546) |
| 0.1 |
2.0 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.1 |
GO:0007521 |
muscle cell fate determination(GO:0007521) |
| 0.1 |
0.8 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 |
0.3 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 |
0.3 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 |
0.1 |
GO:0010046 |
response to mycotoxin(GO:0010046) |
| 0.1 |
0.4 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.2 |
GO:1902396 |
protein localization to bicellular tight junction(GO:1902396) |
| 0.1 |
0.2 |
GO:0046958 |
nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 |
0.3 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.2 |
GO:1902336 |
neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) regulation of retinal ganglion cell axon guidance(GO:0090259) positive regulation of retinal ganglion cell axon guidance(GO:1902336) |
| 0.1 |
0.6 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.2 |
GO:0030862 |
regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 |
0.2 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.4 |
GO:0060178 |
regulation of exocyst localization(GO:0060178) |
| 0.1 |
0.9 |
GO:2000480 |
negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.6 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.4 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.1 |
0.2 |
GO:0016095 |
polyprenol catabolic process(GO:0016095) |
| 0.1 |
0.1 |
GO:0010511 |
regulation of phosphatidylinositol biosynthetic process(GO:0010511) negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.2 |
GO:0009182 |
purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) GDP metabolic process(GO:0046710) |
| 0.1 |
0.4 |
GO:0048069 |
eye pigmentation(GO:0048069) |
| 0.1 |
0.8 |
GO:0050951 |
sensory perception of temperature stimulus(GO:0050951) |
| 0.1 |
0.6 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 |
0.5 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.1 |
0.3 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 |
0.2 |
GO:0007191 |
adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 |
0.3 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.1 |
0.7 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.1 |
0.2 |
GO:0001193 |
maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.1 |
0.8 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.1 |
GO:0021636 |
trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) |
| 0.1 |
0.9 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.2 |
GO:0016598 |
protein arginylation(GO:0016598) |
| 0.1 |
0.4 |
GO:0033299 |
secretion of lysosomal enzymes(GO:0033299) |
| 0.1 |
0.2 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 |
0.3 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.1 |
0.6 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 |
0.3 |
GO:0006382 |
adenosine to inosine editing(GO:0006382) |
| 0.1 |
1.1 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 |
0.2 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.1 |
0.5 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 |
0.2 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.1 |
1.1 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 |
0.2 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.1 |
0.2 |
GO:0038165 |
oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 |
0.4 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.2 |
GO:1901491 |
negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 |
0.2 |
GO:0060708 |
spongiotrophoblast differentiation(GO:0060708) |
| 0.1 |
0.4 |
GO:0016480 |
negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.1 |
0.2 |
GO:2001286 |
regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.1 |
0.5 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.5 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.1 |
0.2 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.1 |
0.5 |
GO:0006012 |
galactose metabolic process(GO:0006012) |
| 0.1 |
0.1 |
GO:0010989 |
negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 |
0.6 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.4 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.1 |
0.4 |
GO:2001224 |
positive regulation of neuron migration(GO:2001224) |
| 0.1 |
0.4 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.1 |
0.2 |
GO:0090467 |
lysine transport(GO:0015819) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) amino acid import into cell(GO:1902837) |
| 0.1 |
0.3 |
GO:0000050 |
urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 |
0.2 |
GO:0086069 |
desmosome assembly(GO:0002159) bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 |
0.1 |
GO:0090237 |
serotonin secretion by platelet(GO:0002554) interleukin-3 production(GO:0032632) regulation of arachidonic acid secretion(GO:0090237) |
| 0.0 |
0.5 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.0 |
0.2 |
GO:0019249 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) |
| 0.0 |
0.1 |
GO:0034035 |
purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 |
0.8 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 |
0.6 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.3 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.3 |
GO:0032380 |
regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 |
0.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.1 |
GO:0003289 |
septum primum development(GO:0003284) atrial septum primum morphogenesis(GO:0003289) |
| 0.0 |
0.6 |
GO:0048715 |
negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 |
1.1 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.2 |
GO:0046391 |
5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 |
0.2 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.0 |
0.2 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.0 |
0.3 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 |
0.3 |
GO:0030539 |
male genitalia development(GO:0030539) |
| 0.0 |
1.2 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 |
0.3 |
GO:0002051 |
osteoblast fate commitment(GO:0002051) |
| 0.0 |
0.1 |
GO:0021898 |
commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 |
0.2 |
GO:0071499 |
response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 |
0.0 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 |
0.2 |
GO:0031077 |
post-embryonic camera-type eye development(GO:0031077) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 |
0.1 |
GO:0035973 |
aggrephagy(GO:0035973) |
| 0.0 |
0.3 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 |
0.1 |
GO:0014834 |
skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 |
0.4 |
GO:0098532 |
histone H3-K27 trimethylation(GO:0098532) |
| 0.0 |
0.2 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.3 |
GO:0071880 |
adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 |
0.7 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.0 |
0.0 |
GO:0048675 |
axon extension(GO:0048675) |
| 0.0 |
0.1 |
GO:1901339 |
regulation of store-operated calcium channel activity(GO:1901339) |
| 0.0 |
0.1 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 |
0.4 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
0.1 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 |
0.3 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 |
0.1 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.0 |
0.1 |
GO:1904529 |
regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.0 |
0.3 |
GO:0090084 |
negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 |
1.4 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
0.5 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.0 |
0.2 |
GO:0010273 |
detoxification of copper ion(GO:0010273) detoxification of inorganic compound(GO:0061687) stress response to copper ion(GO:1990169) |
| 0.0 |
0.4 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 |
0.2 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.0 |
0.1 |
GO:1900533 |
medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 |
0.3 |
GO:0006048 |
UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 |
0.2 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
0.2 |
GO:0006498 |
N-terminal protein lipidation(GO:0006498) |
| 0.0 |
0.3 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.0 |
0.2 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
0.2 |
GO:0035459 |
cargo loading into vesicle(GO:0035459) |
| 0.0 |
0.2 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.0 |
0.1 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 |
0.7 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 |
0.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.6 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 |
0.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.0 |
0.2 |
GO:0046600 |
negative regulation of centriole replication(GO:0046600) |
| 0.0 |
1.3 |
GO:0021884 |
forebrain neuron development(GO:0021884) |
| 0.0 |
0.4 |
GO:0006044 |
N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 |
0.3 |
GO:0019433 |
triglyceride catabolic process(GO:0019433) |
| 0.0 |
0.2 |
GO:0097094 |
craniofacial suture morphogenesis(GO:0097094) |
| 0.0 |
0.2 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.7 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
1.2 |
GO:0001676 |
long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 |
0.1 |
GO:0036092 |
phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 |
0.2 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 |
0.3 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.1 |
GO:1904425 |
negative regulation of GTP binding(GO:1904425) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.4 |
GO:0071549 |
cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 |
0.2 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 |
0.1 |
GO:0071051 |
polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 |
0.2 |
GO:0050765 |
negative regulation of phagocytosis(GO:0050765) |
| 0.0 |
0.2 |
GO:0032957 |
inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 |
0.3 |
GO:0006098 |
pentose-phosphate shunt(GO:0006098) |
| 0.0 |
0.2 |
GO:0021819 |
layer formation in cerebral cortex(GO:0021819) |
| 0.0 |
0.1 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 |
0.1 |
GO:2000813 |
actin filament uncapping(GO:0051695) negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 |
0.2 |
GO:0048733 |
sebaceous gland development(GO:0048733) |
| 0.0 |
0.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.0 |
GO:0097168 |
mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 |
0.4 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.6 |
GO:0001964 |
startle response(GO:0001964) |
| 0.0 |
0.1 |
GO:0045041 |
protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 |
1.6 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.1 |
GO:0006556 |
S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 |
0.2 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 |
0.0 |
GO:0033088 |
negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 |
0.1 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.0 |
0.2 |
GO:0019377 |
glycolipid catabolic process(GO:0019377) |
| 0.0 |
0.1 |
GO:0010748 |
regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 |
0.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.0 |
0.1 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.0 |
0.1 |
GO:1904995 |
negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 |
0.1 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.2 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0099515 |
actin filament-based transport(GO:0099515) |
| 0.0 |
1.8 |
GO:0050709 |
negative regulation of protein secretion(GO:0050709) |
| 0.0 |
1.3 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 |
0.2 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.7 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.2 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.3 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.2 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.1 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 |
0.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.0 |
0.2 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.1 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.1 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.0 |
0.1 |
GO:0008228 |
opsonization(GO:0008228) |
| 0.0 |
0.1 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.1 |
GO:0043691 |
reverse cholesterol transport(GO:0043691) |
| 0.0 |
0.7 |
GO:0045668 |
negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 |
0.1 |
GO:1901387 |
positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 |
0.1 |
GO:0006543 |
glutamine catabolic process(GO:0006543) |
| 0.0 |
0.2 |
GO:0017157 |
regulation of exocytosis(GO:0017157) |
| 0.0 |
0.1 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 |
0.1 |
GO:0040031 |
snRNA modification(GO:0040031) |
| 0.0 |
0.1 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 |
0.0 |
GO:0046726 |
positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 |
0.0 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
0.2 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.1 |
GO:0035927 |
RNA import into mitochondrion(GO:0035927) |
| 0.0 |
0.0 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.0 |
0.3 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
0.1 |
GO:0014886 |
transition between slow and fast fiber(GO:0014886) |
| 0.0 |
0.2 |
GO:0009083 |
branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 |
0.2 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.1 |
GO:0035795 |
negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.0 |
0.1 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 |
0.4 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.1 |
GO:1903799 |
negative regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903799) |
| 0.0 |
0.3 |
GO:0006904 |
vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 |
0.2 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
0.0 |
GO:0006434 |
seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 |
0.5 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.4 |
GO:0001954 |
positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 |
0.1 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 |
0.2 |
GO:0042407 |
cristae formation(GO:0042407) |
| 0.0 |
0.1 |
GO:0000059 |
protein import into nucleus, docking(GO:0000059) |
| 0.0 |
0.2 |
GO:0043496 |
regulation of protein homodimerization activity(GO:0043496) |
| 0.0 |
0.3 |
GO:0048148 |
behavioral response to cocaine(GO:0048148) |
| 0.0 |
0.1 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 |
0.4 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.2 |
GO:0044380 |
protein localization to cytoskeleton(GO:0044380) |
| 0.0 |
0.0 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 |
0.0 |
GO:0060029 |
convergent extension involved in organogenesis(GO:0060029) |
| 0.0 |
1.1 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.0 |
0.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.0 |
0.1 |
GO:0045740 |
positive regulation of DNA replication(GO:0045740) |
| 0.0 |
0.5 |
GO:0007416 |
synapse assembly(GO:0007416) |
| 0.0 |
0.7 |
GO:0032411 |
positive regulation of transporter activity(GO:0032411) |
| 0.0 |
0.1 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.0 |
0.1 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.0 |
GO:0010387 |
COP9 signalosome assembly(GO:0010387) |
| 0.0 |
0.4 |
GO:0021680 |
cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 |
0.3 |
GO:0002931 |
response to ischemia(GO:0002931) |
| 0.0 |
0.9 |
GO:0030641 |
regulation of cellular pH(GO:0030641) |
| 0.0 |
0.0 |
GO:0048680 |
positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 |
0.4 |
GO:0010812 |
negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 |
0.0 |
GO:0035964 |
COPI-coated vesicle budding(GO:0035964) |
| 0.0 |
0.2 |
GO:0033003 |
regulation of mast cell activation(GO:0033003) |
| 0.0 |
0.2 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.1 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.2 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
0.0 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.1 |
GO:0090083 |
regulation of inclusion body assembly(GO:0090083) |
| 0.0 |
0.2 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.3 |
GO:0045746 |
negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 |
0.2 |
GO:0048168 |
regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 |
0.1 |
GO:0060074 |
synapse maturation(GO:0060074) |