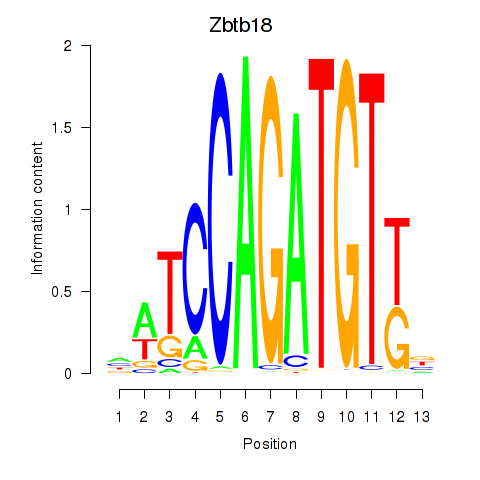
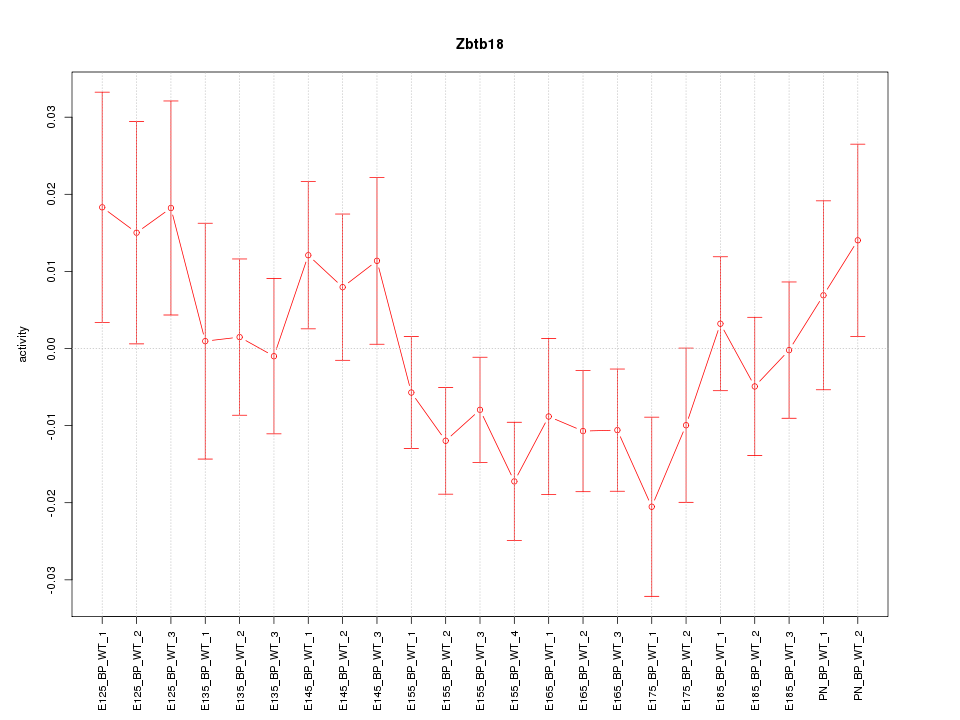
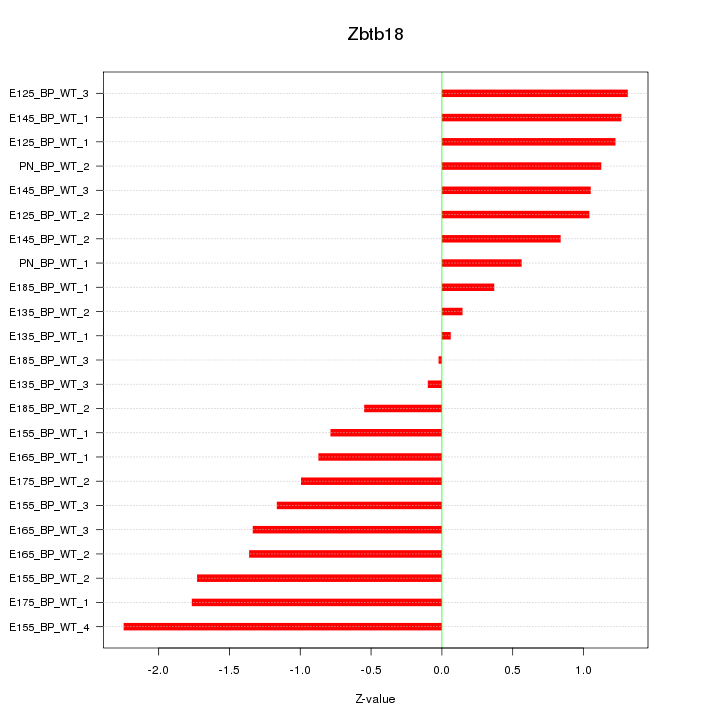
| 1.7 |
5.1 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.7 |
2.0 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.5 |
3.3 |
GO:0016576 |
histone dephosphorylation(GO:0016576) |
| 0.4 |
1.3 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.4 |
1.1 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.3 |
1.0 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 |
1.0 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.3 |
1.2 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.3 |
0.9 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.3 |
0.8 |
GO:1901421 |
positive regulation of response to alcohol(GO:1901421) |
| 0.3 |
1.3 |
GO:0070327 |
thyroid hormone transport(GO:0070327) |
| 0.2 |
0.7 |
GO:0010757 |
arachidonic acid metabolite production involved in inflammatory response(GO:0002538) negative regulation of plasminogen activation(GO:0010757) regulation of vascular wound healing(GO:0061043) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 |
1.0 |
GO:0060282 |
positive regulation of oocyte development(GO:0060282) |
| 0.2 |
2.1 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 |
1.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.2 |
0.6 |
GO:0002940 |
tRNA N2-guanine methylation(GO:0002940) |
| 0.2 |
5.1 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 |
2.5 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.2 |
0.6 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.2 |
2.0 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.2 |
3.7 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.2 |
0.9 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 |
0.5 |
GO:0015960 |
diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.2 |
1.4 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.2 |
0.9 |
GO:1901837 |
negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 |
0.3 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.2 |
1.2 |
GO:2000857 |
positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 |
0.7 |
GO:0051661 |
maintenance of centrosome location(GO:0051661) |
| 0.1 |
0.6 |
GO:0006529 |
asparagine biosynthetic process(GO:0006529) |
| 0.1 |
0.6 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 |
0.3 |
GO:0002295 |
T-helper cell lineage commitment(GO:0002295) |
| 0.1 |
1.3 |
GO:0044336 |
canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 |
0.9 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.1 |
0.7 |
GO:0097527 |
necroptotic signaling pathway(GO:0097527) |
| 0.1 |
0.6 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.1 |
0.4 |
GO:0032918 |
polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 |
0.7 |
GO:0045163 |
clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 |
0.6 |
GO:0090234 |
cellular response to testosterone stimulus(GO:0071394) regulation of kinetochore assembly(GO:0090234) |
| 0.1 |
1.2 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
0.5 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
1.1 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.3 |
GO:0032488 |
Cdc42 protein signal transduction(GO:0032488) |
| 0.1 |
1.0 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 |
0.3 |
GO:0018094 |
protein polyglycylation(GO:0018094) |
| 0.1 |
0.4 |
GO:0032511 |
late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 |
0.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 0.1 |
0.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 |
0.4 |
GO:0071033 |
nuclear retention of pre-mRNA at the site of transcription(GO:0071033) CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 |
0.9 |
GO:0009186 |
deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.1 |
0.6 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
1.3 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.1 |
3.2 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 |
1.7 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
3.1 |
GO:0014003 |
oligodendrocyte development(GO:0014003) |
| 0.1 |
0.2 |
GO:2000040 |
regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 |
0.2 |
GO:0006597 |
spermine biosynthetic process(GO:0006597) |
| 0.1 |
0.5 |
GO:0032796 |
uropod organization(GO:0032796) |
| 0.1 |
0.7 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.4 |
GO:0007320 |
insemination(GO:0007320) |
| 0.1 |
0.6 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.1 |
1.6 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.1 |
0.3 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 0.1 |
0.6 |
GO:0043117 |
positive regulation of vascular permeability(GO:0043117) |
| 0.1 |
0.2 |
GO:0001803 |
antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.1 |
1.4 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.5 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.1 |
1.1 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.1 |
2.6 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |
| 0.1 |
0.6 |
GO:0010992 |
ubiquitin homeostasis(GO:0010992) |
| 0.1 |
0.3 |
GO:0032532 |
regulation of microvillus length(GO:0032532) |
| 0.1 |
0.2 |
GO:0006547 |
histidine metabolic process(GO:0006547) |
| 0.1 |
0.2 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 |
0.8 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.3 |
GO:2000491 |
positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 |
0.5 |
GO:1900119 |
positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 |
0.5 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.7 |
GO:0046855 |
phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 |
0.3 |
GO:0007128 |
meiotic prophase I(GO:0007128) prophase(GO:0051324) meiotic cell cycle phase(GO:0098762) meiosis I cell cycle phase(GO:0098764) |
| 0.0 |
1.3 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.1 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 |
0.2 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 |
0.2 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) protein localization to adherens junction(GO:0071896) |
| 0.0 |
0.5 |
GO:0043252 |
sodium-independent organic anion transport(GO:0043252) |
| 0.0 |
0.8 |
GO:0006958 |
complement activation, classical pathway(GO:0006958) |
| 0.0 |
0.2 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.7 |
GO:0032288 |
myelin assembly(GO:0032288) |
| 0.0 |
0.4 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.0 |
0.1 |
GO:0040009 |
regulation of growth rate(GO:0040009) protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 |
0.2 |
GO:0006549 |
isoleucine metabolic process(GO:0006549) branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.2 |
GO:0033762 |
response to glucagon(GO:0033762) |
| 0.0 |
0.4 |
GO:0090308 |
regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 0.0 |
0.1 |
GO:0060290 |
transdifferentiation(GO:0060290) |
| 0.0 |
0.4 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
0.1 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) potassium ion export across plasma membrane(GO:0097623) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.0 |
0.1 |
GO:0048539 |
bone marrow development(GO:0048539) |
| 0.0 |
0.6 |
GO:0002076 |
osteoblast development(GO:0002076) |
| 0.0 |
0.1 |
GO:0034472 |
snRNA 3'-end processing(GO:0034472) |
| 0.0 |
0.2 |
GO:1901727 |
positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 |
0.2 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
1.1 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 |
0.1 |
GO:0042732 |
D-xylose metabolic process(GO:0042732) |
| 0.0 |
0.2 |
GO:0070389 |
chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 |
0.3 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 |
0.1 |
GO:0019401 |
alditol biosynthetic process(GO:0019401) |
| 0.0 |
0.2 |
GO:0003065 |
positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 |
0.9 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.1 |
GO:1902037 |
negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 |
4.7 |
GO:0030308 |
negative regulation of cell growth(GO:0030308) |
| 0.0 |
0.6 |
GO:0006491 |
N-glycan processing(GO:0006491) |
| 0.0 |
1.7 |
GO:0007338 |
single fertilization(GO:0007338) |
| 0.0 |
1.4 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.2 |
GO:0006370 |
7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 |
0.1 |
GO:0006451 |
selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 |
0.1 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.0 |
0.2 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 |
0.1 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 |
0.7 |
GO:0006801 |
superoxide metabolic process(GO:0006801) |
| 0.0 |
0.3 |
GO:0046628 |
positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 |
0.2 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.0 |
0.9 |
GO:0042771 |
intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 |
0.1 |
GO:0097278 |
virion attachment to host cell(GO:0019062) adhesion of symbiont to host cell(GO:0044650) complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.2 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.2 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.0 |
0.1 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0019659 |
glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 |
0.4 |
GO:0000956 |
nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 |
1.2 |
GO:0007093 |
mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.1 |
GO:0009136 |
ADP biosynthetic process(GO:0006172) nucleoside diphosphate biosynthetic process(GO:0009133) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 |
0.1 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.0 |
0.3 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 |
0.1 |
GO:0034498 |
early endosome to Golgi transport(GO:0034498) |
| 0.0 |
0.2 |
GO:0046685 |
response to arsenic-containing substance(GO:0046685) |
| 0.0 |
0.1 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
1.0 |
GO:0034620 |
cellular response to unfolded protein(GO:0034620) |
| 0.0 |
0.8 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
1.3 |
GO:0000086 |
G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 |
0.4 |
GO:0048678 |
response to axon injury(GO:0048678) |
| 0.0 |
0.3 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.1 |
GO:0035814 |
negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 |
0.5 |
GO:0071346 |
cellular response to interferon-gamma(GO:0071346) |
| 0.0 |
0.5 |
GO:0001938 |
positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 |
0.3 |
GO:0006783 |
heme biosynthetic process(GO:0006783) |
| 0.0 |
0.3 |
GO:0032008 |
positive regulation of TOR signaling(GO:0032008) |
| 0.0 |
0.1 |
GO:0070813 |
hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 |
0.3 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.3 |
GO:0033209 |
tumor necrosis factor-mediated signaling pathway(GO:0033209) |