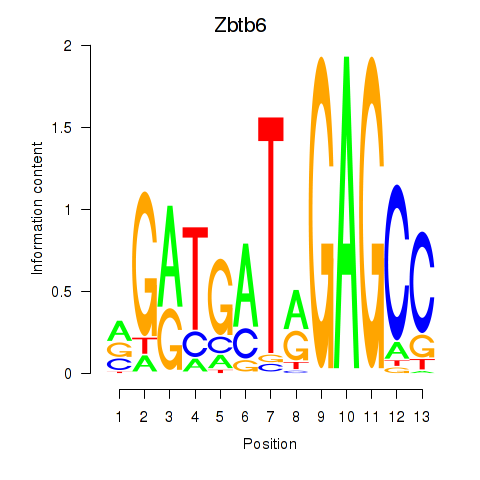
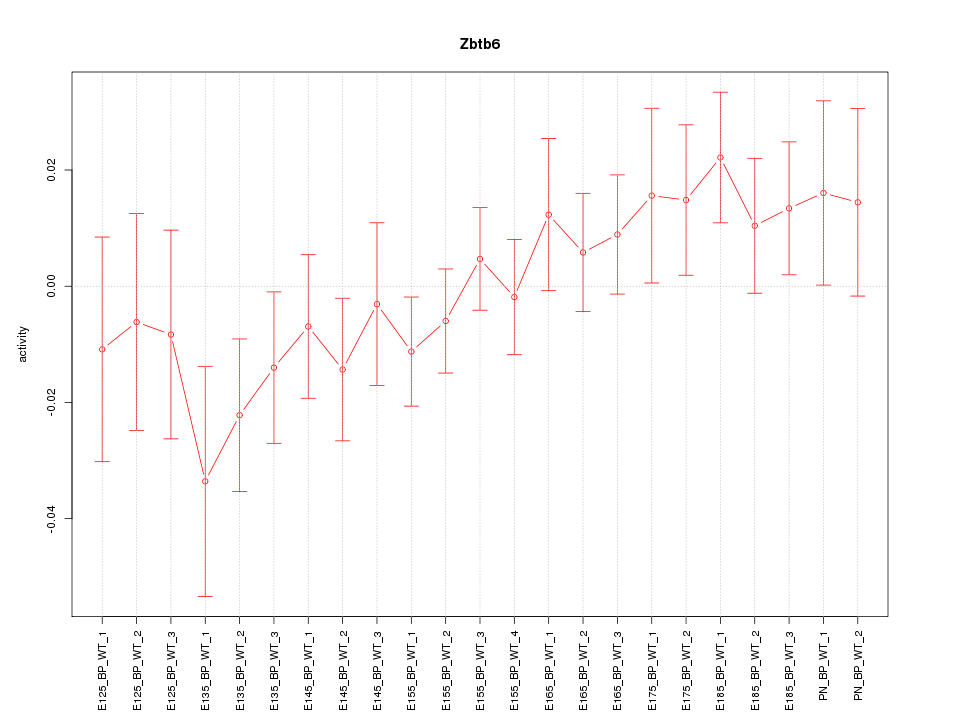
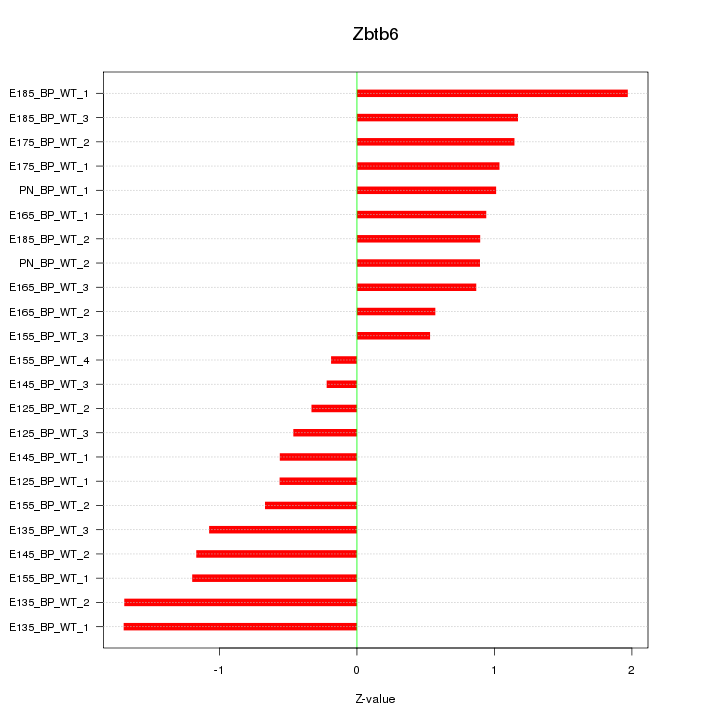
| 1.6 |
4.9 |
GO:0021837 |
motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 1.6 |
9.8 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) |
| 1.6 |
4.7 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 0.8 |
3.1 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.7 |
5.0 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.6 |
1.8 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.6 |
1.7 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.5 |
1.4 |
GO:2000564 |
CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.5 |
1.4 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) transepithelial ammonium transport(GO:0070634) |
| 0.4 |
4.2 |
GO:2001214 |
positive regulation of vasculogenesis(GO:2001214) |
| 0.4 |
2.1 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.3 |
3.4 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.3 |
3.3 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.3 |
6.4 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.3 |
0.8 |
GO:0003278 |
apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.3 |
1.9 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 |
3.8 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 |
1.1 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 |
0.6 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.2 |
0.8 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.2 |
0.5 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.1 |
0.9 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 |
0.4 |
GO:0036500 |
ATF6-mediated unfolded protein response(GO:0036500) |
| 0.1 |
0.8 |
GO:1903912 |
negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 |
2.0 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.7 |
GO:1903361 |
protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 |
0.4 |
GO:0050929 |
corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.1 |
0.7 |
GO:0036515 |
serotonergic neuron axon guidance(GO:0036515) |
| 0.1 |
0.3 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 |
0.5 |
GO:0034975 |
protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 |
0.6 |
GO:0015803 |
branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 |
0.9 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.1 |
0.5 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.1 |
4.3 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.1 |
0.7 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 |
0.3 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.1 |
0.3 |
GO:0048294 |
negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 |
0.2 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 0.1 |
1.2 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
0.6 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
0.5 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 |
0.2 |
GO:0043201 |
response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 |
1.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.1 |
1.5 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.5 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 |
1.5 |
GO:0030318 |
melanocyte differentiation(GO:0030318) |
| 0.0 |
1.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.1 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) |
| 0.0 |
1.7 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
0.3 |
GO:0048840 |
otolith development(GO:0048840) |
| 0.0 |
0.6 |
GO:0006555 |
methionine metabolic process(GO:0006555) |
| 0.0 |
0.5 |
GO:0031937 |
positive regulation of chromatin silencing(GO:0031937) |
| 0.0 |
1.0 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.0 |
0.3 |
GO:0071318 |
cellular response to ATP(GO:0071318) |
| 0.0 |
0.1 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.1 |
GO:0032916 |
positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 |
0.3 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.8 |
GO:0046854 |
phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 |
0.7 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.3 |
GO:0046415 |
urate metabolic process(GO:0046415) |
| 0.0 |
0.7 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.1 |
GO:2000347 |
positive regulation of hepatocyte proliferation(GO:2000347) |
| 0.0 |
0.2 |
GO:0097151 |
spontaneous neurotransmitter secretion(GO:0061669) positive regulation of inhibitory postsynaptic potential(GO:0097151) spontaneous synaptic transmission(GO:0098814) |
| 0.0 |
0.2 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.5 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.3 |
GO:0044243 |
collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.9 |
GO:0010718 |
positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 |
0.6 |
GO:1904030 |
negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 |
0.1 |
GO:0043506 |
regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 |
0.4 |
GO:0045601 |
regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 |
0.2 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.1 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.0 |
0.5 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.0 |
0.7 |
GO:0006893 |
Golgi to plasma membrane transport(GO:0006893) |
| 0.0 |
0.4 |
GO:0030199 |
collagen fibril organization(GO:0030199) |
| 0.0 |
0.4 |
GO:0086010 |
membrane depolarization during action potential(GO:0086010) |