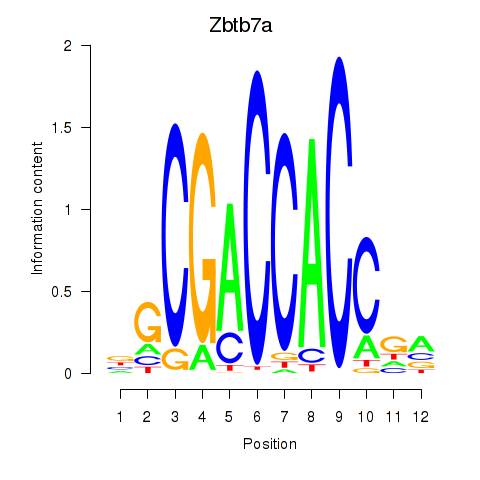
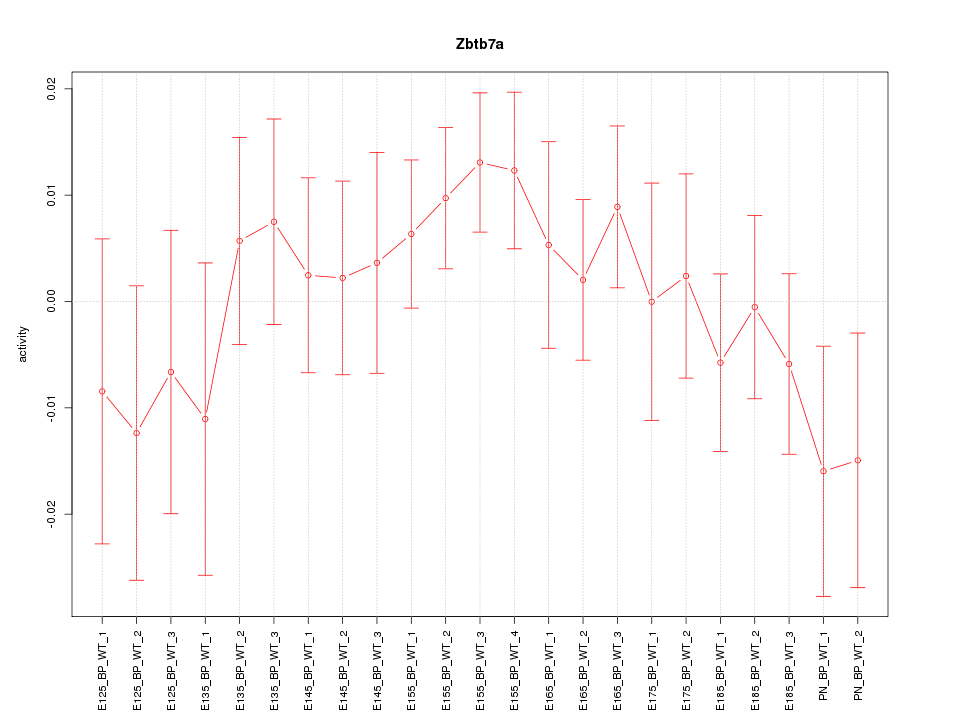
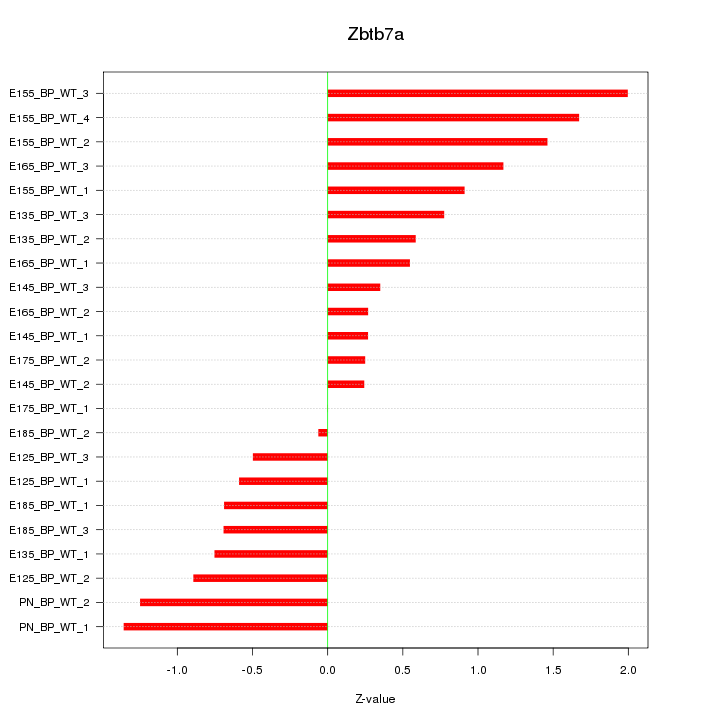
| 0.7 |
2.9 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.5 |
1.9 |
GO:0060912 |
cardiac cell fate specification(GO:0060912) |
| 0.4 |
1.8 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 |
1.1 |
GO:0050975 |
sensory perception of touch(GO:0050975) |
| 0.3 |
1.0 |
GO:0003134 |
BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.3 |
0.9 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.3 |
0.9 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.3 |
1.1 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.3 |
1.4 |
GO:0001778 |
plasma membrane repair(GO:0001778) |
| 0.3 |
1.4 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.3 |
0.8 |
GO:1904742 |
protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.3 |
0.8 |
GO:0097401 |
synaptic vesicle lumen acidification(GO:0097401) |
| 0.3 |
1.3 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.2 |
1.5 |
GO:1902837 |
amino acid import into cell(GO:1902837) |
| 0.2 |
0.7 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.2 |
0.9 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
0.9 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.2 |
0.6 |
GO:0071336 |
lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.2 |
1.2 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
0.6 |
GO:2000170 |
positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 |
0.8 |
GO:0009597 |
detection of virus(GO:0009597) |
| 0.2 |
0.6 |
GO:0002035 |
brain renin-angiotensin system(GO:0002035) |
| 0.2 |
0.7 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.2 |
1.0 |
GO:1901524 |
regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 0.2 |
0.8 |
GO:0034436 |
glycoprotein transport(GO:0034436) |
| 0.1 |
0.6 |
GO:0045343 |
MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.1 |
0.4 |
GO:0051387 |
negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 |
0.5 |
GO:0019355 |
nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 |
1.0 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.4 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 |
0.5 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.5 |
GO:2000054 |
negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 |
0.5 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.1 |
0.3 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
1.0 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
0.3 |
GO:0016256 |
N-glycan processing to lysosome(GO:0016256) |
| 0.1 |
1.2 |
GO:0070970 |
interleukin-2 secretion(GO:0070970) |
| 0.1 |
0.3 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 0.1 |
0.5 |
GO:0007223 |
muscular septum morphogenesis(GO:0003150) Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 |
1.4 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 |
0.3 |
GO:0014891 |
striated muscle atrophy(GO:0014891) |
| 0.1 |
0.8 |
GO:0002903 |
negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 |
0.4 |
GO:0002536 |
respiratory burst involved in inflammatory response(GO:0002536) |
| 0.1 |
0.5 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.1 |
0.5 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 |
0.2 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 |
0.7 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 |
0.3 |
GO:1900740 |
regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.1 |
0.2 |
GO:0032058 |
positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 |
0.2 |
GO:2000744 |
anterior head development(GO:0097065) regulation of anterior head development(GO:2000742) positive regulation of anterior head development(GO:2000744) |
| 0.1 |
0.4 |
GO:0045078 |
positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.1 |
2.0 |
GO:0042059 |
negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) |
| 0.1 |
0.2 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 |
0.6 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.5 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.7 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.1 |
0.8 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.8 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 |
0.3 |
GO:0048631 |
regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.1 |
0.2 |
GO:0072070 |
loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.1 |
0.3 |
GO:0071947 |
protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 |
0.2 |
GO:0072718 |
response to cisplatin(GO:0072718) |
| 0.1 |
0.4 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.1 |
0.2 |
GO:0070164 |
negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 |
0.2 |
GO:0046061 |
dATP catabolic process(GO:0046061) |
| 0.1 |
0.2 |
GO:0055099 |
response to high density lipoprotein particle(GO:0055099) |
| 0.1 |
0.7 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.2 |
GO:0032240 |
negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.1 |
0.6 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.1 |
0.8 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.1 |
0.3 |
GO:0098838 |
reduced folate transmembrane transport(GO:0098838) |
| 0.1 |
0.2 |
GO:0032650 |
regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) interleukin-1 alpha secretion(GO:0050703) |
| 0.0 |
0.7 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.4 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 |
0.2 |
GO:0033700 |
phospholipid efflux(GO:0033700) |
| 0.0 |
0.6 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.0 |
0.4 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
1.0 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.0 |
0.3 |
GO:0060527 |
prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.0 |
0.6 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 |
0.3 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.0 |
0.4 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.0 |
0.2 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.0 |
0.2 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.1 |
GO:0032532 |
regulation of microvillus length(GO:0032532) |
| 0.0 |
0.1 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.2 |
GO:0001302 |
replicative cell aging(GO:0001302) |
| 0.0 |
0.2 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 |
0.1 |
GO:0097211 |
response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 |
0.4 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.0 |
0.1 |
GO:1904008 |
response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.0 |
0.6 |
GO:0000188 |
inactivation of MAPK activity(GO:0000188) |
| 0.0 |
0.8 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.3 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.0 |
0.2 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.6 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.1 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.0 |
0.2 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 |
1.5 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.0 |
0.3 |
GO:0070445 |
oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 |
0.2 |
GO:1900121 |
negative regulation of receptor binding(GO:1900121) |
| 0.0 |
0.1 |
GO:0010940 |
positive regulation of necrotic cell death(GO:0010940) |
| 0.0 |
0.2 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.1 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.1 |
GO:0018364 |
peptidyl-glutamine methylation(GO:0018364) |
| 0.0 |
0.1 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 |
0.3 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 0.0 |
0.3 |
GO:0046835 |
carbohydrate phosphorylation(GO:0046835) |
| 0.0 |
0.1 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 |
0.4 |
GO:0010592 |
positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 |
0.2 |
GO:0090385 |
phagosome-lysosome fusion(GO:0090385) |
| 0.0 |
0.1 |
GO:0051790 |
short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 |
0.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.0 |
GO:0009051 |
pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 |
0.2 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0061052 |
negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 |
0.0 |
GO:0061156 |
pulmonary artery morphogenesis(GO:0061156) |
| 0.0 |
0.1 |
GO:0014835 |
myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 |
0.3 |
GO:0061339 |
establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 |
0.3 |
GO:0060055 |
angiogenesis involved in wound healing(GO:0060055) |
| 0.0 |
0.1 |
GO:0007253 |
cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 |
0.7 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.1 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.0 |
0.1 |
GO:0003383 |
apical constriction(GO:0003383) |
| 0.0 |
0.1 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 |
1.5 |
GO:0051897 |
positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 |
0.1 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 |
0.3 |
GO:0014898 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.2 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.3 |
GO:0036010 |
protein localization to endosome(GO:0036010) |
| 0.0 |
0.1 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 |
0.1 |
GO:0043137 |
DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 |
0.3 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.0 |
0.3 |
GO:0035067 |
negative regulation of histone acetylation(GO:0035067) |
| 0.0 |
0.5 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.2 |
GO:0071624 |
positive regulation of granulocyte chemotaxis(GO:0071624) positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 |
0.3 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 |
0.3 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.0 |
GO:0015684 |
ferrous iron transport(GO:0015684) |
| 0.0 |
0.1 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
0.3 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.3 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 |
0.2 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |
| 0.0 |
0.2 |
GO:0006264 |
mitochondrial DNA replication(GO:0006264) |
| 0.0 |
0.6 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 |
0.2 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.2 |
GO:0045943 |
positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 |
0.2 |
GO:0090005 |
negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 |
0.1 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.0 |
0.1 |
GO:0034427 |
nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 |
0.2 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 |
0.1 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
0.4 |
GO:0051881 |
regulation of mitochondrial membrane potential(GO:0051881) |
| 0.0 |
0.2 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 |
0.0 |
GO:0002339 |
B cell selection(GO:0002339) |