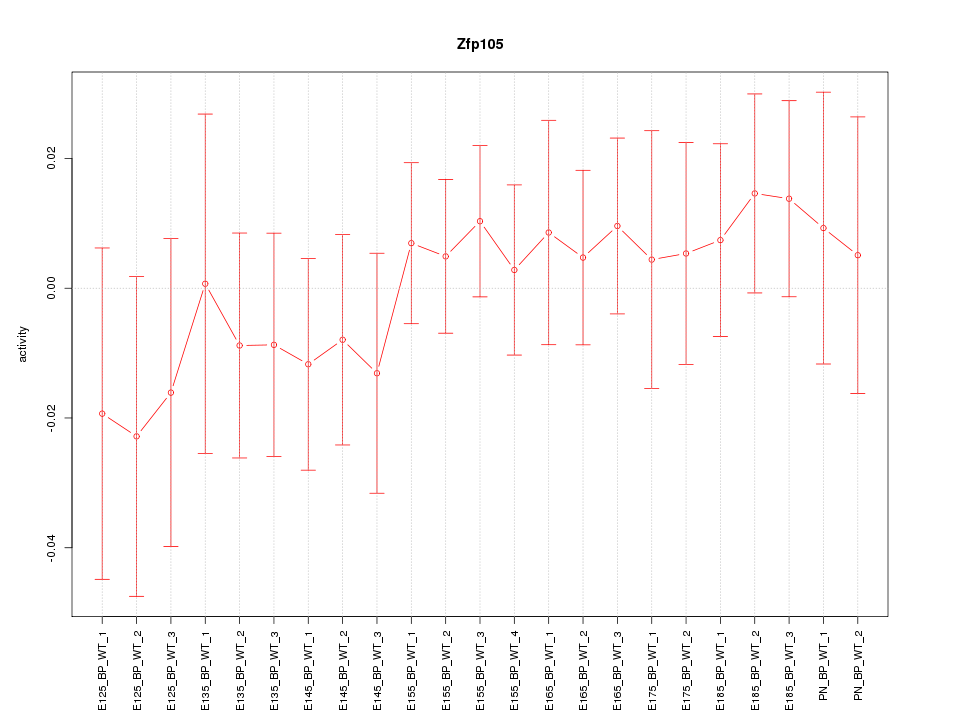
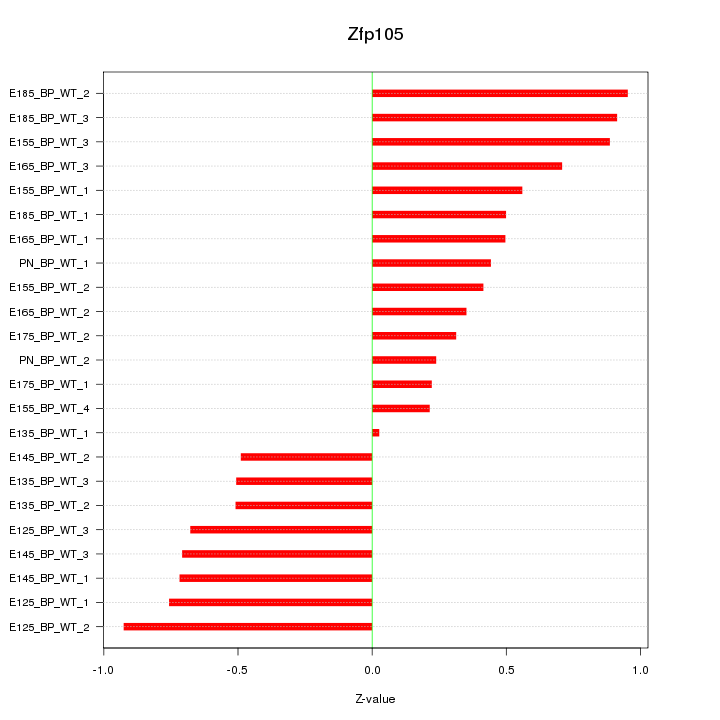
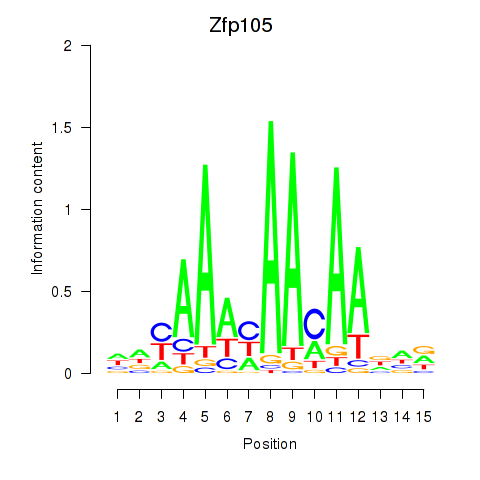
Motif ID: Zfp105
Z-value: 0.599
Transcription factors associated with Zfp105:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Zfp105 | ENSMUSG00000057895.5 | Zfp105 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp105 | mm10_v2_chr9_+_122923050_122923099 | 0.35 | 9.9e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.7 | 2.0 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.5 | 5.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 0.9 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 1.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.2 | 2.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.6 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 1.5 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.1 | 0.5 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.3 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.1 | 0.5 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.5 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 1.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.2 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.5 | 5.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.5 | 2.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.4 | 1.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.3 | 2.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.5 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 0.4 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |