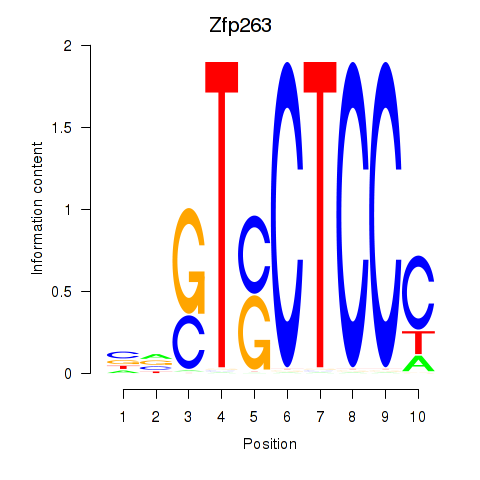
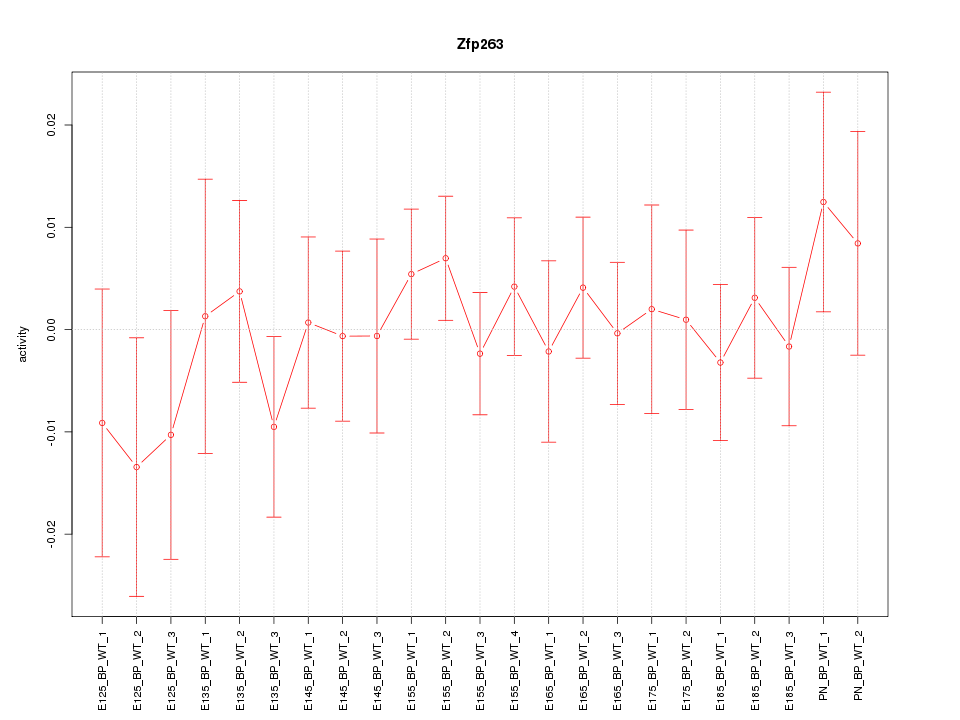
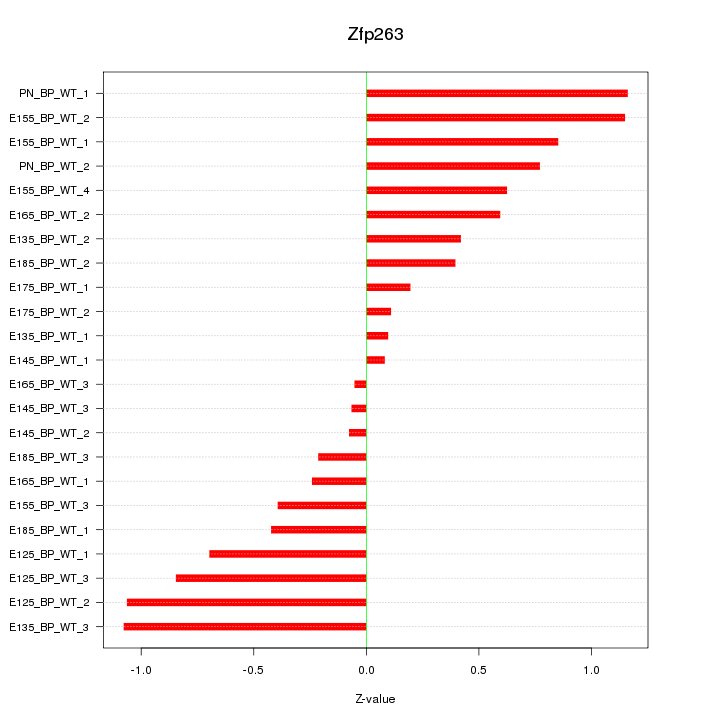
| 1.1 |
3.3 |
GO:0072223 |
metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.5 |
1.6 |
GO:0043379 |
memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.5 |
2.0 |
GO:1903416 |
response to glycoside(GO:1903416) |
| 0.4 |
1.7 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.4 |
1.2 |
GO:1904956 |
dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.3 |
1.0 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.3 |
1.2 |
GO:0003430 |
growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 |
1.2 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.3 |
0.9 |
GO:0006001 |
fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 |
0.9 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.3 |
0.8 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.3 |
1.1 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.3 |
0.5 |
GO:0099624 |
regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) atrial cardiac muscle cell membrane repolarization(GO:0099624) |
| 0.2 |
0.7 |
GO:0035621 |
ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.2 |
0.6 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.2 |
0.6 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 |
0.6 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.2 |
2.8 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 |
0.4 |
GO:0032829 |
tolerance induction to self antigen(GO:0002513) regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.2 |
0.6 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 |
0.5 |
GO:0003289 |
atrial septum primum morphogenesis(GO:0003289) |
| 0.2 |
1.2 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 |
0.1 |
GO:0003308 |
negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 |
0.4 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 |
0.7 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.6 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.9 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.1 |
0.4 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.1 |
0.6 |
GO:0021914 |
negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 |
0.2 |
GO:0042663 |
regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 |
0.4 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 |
0.6 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.1 |
0.7 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
0.6 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.8 |
GO:0015670 |
carbon dioxide transport(GO:0015670) |
| 0.1 |
0.4 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 |
0.6 |
GO:0070100 |
negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 |
1.0 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
0.4 |
GO:0061010 |
gall bladder development(GO:0061010) |
| 0.1 |
0.3 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 0.1 |
0.3 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.3 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 |
0.2 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 |
0.3 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 |
0.5 |
GO:0034441 |
plasma lipoprotein particle oxidation(GO:0034441) |
| 0.1 |
0.6 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 |
0.6 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.3 |
GO:0051599 |
response to hydrostatic pressure(GO:0051599) |
| 0.1 |
1.3 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 |
0.5 |
GO:0051541 |
elastin metabolic process(GO:0051541) |
| 0.1 |
0.4 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 |
1.7 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.4 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 |
0.6 |
GO:0072602 |
interleukin-4 secretion(GO:0072602) |
| 0.1 |
0.3 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 |
0.2 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.1 |
0.4 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
2.3 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.1 |
0.2 |
GO:0003134 |
heart induction(GO:0003129) BMP signaling pathway involved in heart induction(GO:0003130) endodermal-mesodermal cell signaling(GO:0003133) endodermal-mesodermal cell signaling involved in heart induction(GO:0003134) nephric duct formation(GO:0072179) |
| 0.1 |
1.0 |
GO:0043252 |
prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.1 |
0.4 |
GO:1900246 |
positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.1 |
0.3 |
GO:0046469 |
platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.1 |
0.2 |
GO:0051342 |
regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 |
0.2 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 |
0.3 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.2 |
GO:0061030 |
ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 |
0.7 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
0.9 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.1 |
0.3 |
GO:0014846 |
esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 |
0.2 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 |
0.3 |
GO:0006072 |
glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 |
0.2 |
GO:0060715 |
syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.1 |
0.2 |
GO:0006049 |
UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 |
0.6 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 |
0.2 |
GO:0090069 |
regulation of ribosome biogenesis(GO:0090069) |
| 0.1 |
0.4 |
GO:2000258 |
negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 |
0.9 |
GO:0014051 |
gamma-aminobutyric acid secretion(GO:0014051) |
| 0.1 |
0.2 |
GO:0006295 |
pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) positive regulation of t-circle formation(GO:1904431) |
| 0.1 |
0.5 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 |
0.3 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.4 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.0 |
0.3 |
GO:1901660 |
calcium ion export(GO:1901660) |
| 0.0 |
0.1 |
GO:2000275 |
regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 |
0.1 |
GO:0045014 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.0 |
0.3 |
GO:1990118 |
sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 |
0.2 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.2 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.0 |
0.7 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 |
0.4 |
GO:0051608 |
histamine transport(GO:0051608) |
| 0.0 |
0.1 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 |
0.3 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 |
2.5 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.0 |
0.1 |
GO:0097309 |
cap1 mRNA methylation(GO:0097309) |
| 0.0 |
0.2 |
GO:0016081 |
synaptic vesicle docking(GO:0016081) |
| 0.0 |
1.3 |
GO:0009950 |
dorsal/ventral axis specification(GO:0009950) |
| 0.0 |
0.2 |
GO:0042795 |
snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 |
1.2 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.0 |
0.1 |
GO:0050748 |
negative regulation of receptor biosynthetic process(GO:0010871) negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.0 |
0.4 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 |
0.2 |
GO:1903587 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.0 |
0.3 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 |
0.3 |
GO:0035898 |
parathyroid hormone secretion(GO:0035898) |
| 0.0 |
0.6 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 |
0.3 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 |
0.4 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.2 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 0.0 |
0.4 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.3 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.0 |
0.3 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.0 |
0.2 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 |
0.7 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.0 |
0.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 |
0.2 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 |
0.1 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 |
0.3 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.8 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
0.2 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 |
0.3 |
GO:0000463 |
maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 |
0.1 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.0 |
0.3 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.0 |
0.1 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.2 |
GO:0048168 |
regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 |
0.2 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 |
0.4 |
GO:0031340 |
positive regulation of vesicle fusion(GO:0031340) |
| 0.0 |
0.2 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.2 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 |
0.2 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.2 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.5 |
GO:0007614 |
short-term memory(GO:0007614) |
| 0.0 |
0.2 |
GO:0060903 |
positive regulation of meiosis I(GO:0060903) |
| 0.0 |
0.2 |
GO:0010587 |
miRNA catabolic process(GO:0010587) |
| 0.0 |
0.2 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.0 |
0.1 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 |
0.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 |
0.1 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 |
0.1 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 |
0.1 |
GO:2000271 |
positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 |
0.1 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.0 |
0.1 |
GO:0031033 |
myosin filament organization(GO:0031033) |
| 0.0 |
0.7 |
GO:0045670 |
regulation of osteoclast differentiation(GO:0045670) |
| 0.0 |
0.2 |
GO:0099515 |
actin filament-based transport(GO:0099515) |
| 0.0 |
0.2 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.7 |
GO:0070206 |
protein trimerization(GO:0070206) |
| 0.0 |
0.0 |
GO:0034392 |
negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 |
0.1 |
GO:0045651 |
positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 |
0.2 |
GO:0060914 |
heart formation(GO:0060914) |
| 0.0 |
0.3 |
GO:0014912 |
negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 |
0.0 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.0 |
0.1 |
GO:1901998 |
toxin transport(GO:1901998) |
| 0.0 |
0.1 |
GO:0061179 |
negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 |
0.2 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
0.1 |
GO:0051924 |
regulation of calcium ion transport(GO:0051924) |
| 0.0 |
0.9 |
GO:0016525 |
negative regulation of angiogenesis(GO:0016525) |
| 0.0 |
0.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.8 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.1 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.1 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 |
0.2 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.0 |
GO:0002921 |
negative regulation of humoral immune response(GO:0002921) |
| 0.0 |
0.1 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.0 |
0.4 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.0 |
0.0 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.0 |
1.0 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:0051096 |
telomere assembly(GO:0032202) regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 |
0.2 |
GO:0032291 |
central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 |
0.8 |
GO:0051592 |
response to calcium ion(GO:0051592) |
| 0.0 |
0.1 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 |
0.1 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 |
0.1 |
GO:0048715 |
negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 |
0.7 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
2.0 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.1 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.0 |
0.0 |
GO:0061009 |
common bile duct development(GO:0061009) |