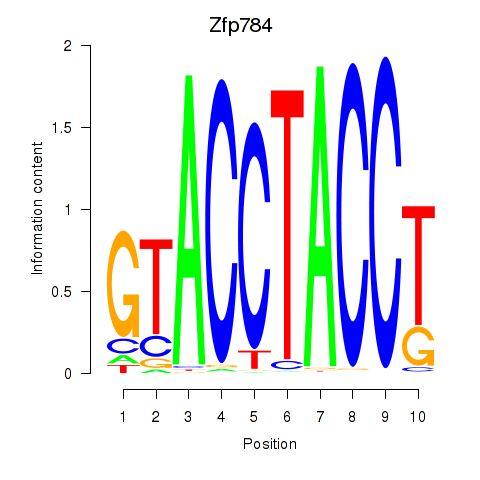
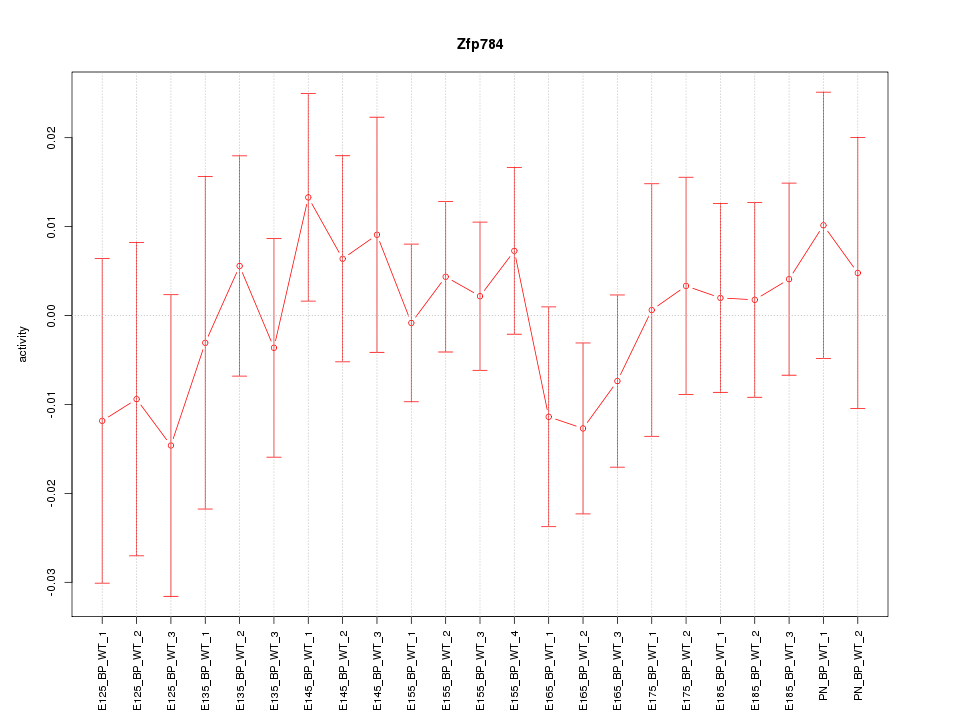
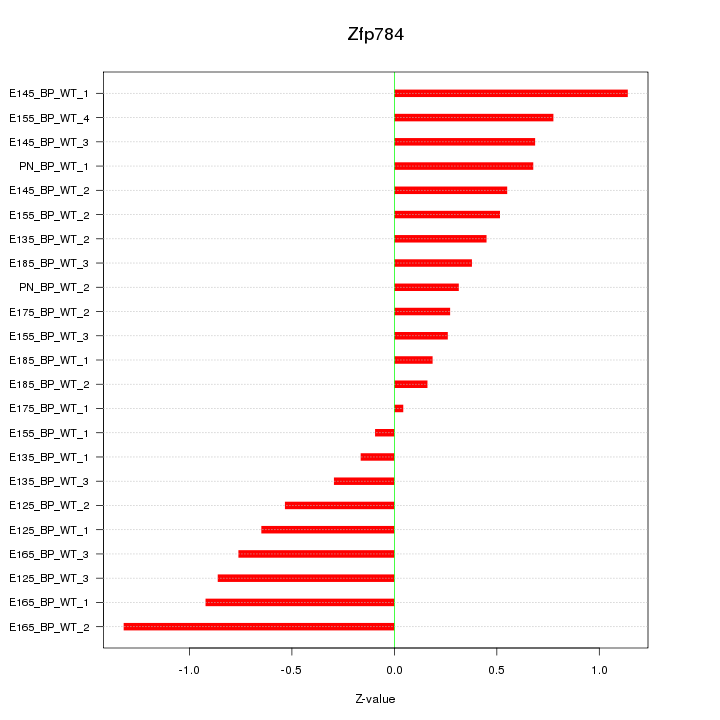
| 0.7 |
2.0 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.5 |
1.5 |
GO:2000097 |
regulation of smooth muscle cell-matrix adhesion(GO:2000097) negative regulation of lymphocyte migration(GO:2000402) |
| 0.4 |
1.3 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.3 |
2.0 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 |
1.3 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 |
0.9 |
GO:0035441 |
cell migration involved in vasculogenesis(GO:0035441) metanephric glomerular mesangium development(GO:0072223) metanephric glomerular mesangial cell proliferation involved in metanephros development(GO:0072262) |
| 0.3 |
1.2 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 |
0.8 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.3 |
1.1 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.2 |
1.6 |
GO:0030388 |
fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 |
0.7 |
GO:0061073 |
ciliary body morphogenesis(GO:0061073) |
| 0.1 |
0.5 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 |
0.6 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 |
0.5 |
GO:0017183 |
peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 |
0.5 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.1 |
0.5 |
GO:0032914 |
positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.1 |
0.6 |
GO:0071376 |
response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 |
0.3 |
GO:0045404 |
positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 |
0.3 |
GO:0010751 |
regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 |
0.4 |
GO:2000987 |
positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 |
0.4 |
GO:0071205 |
regulation of axon diameter(GO:0031133) clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.1 |
0.6 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.1 |
0.7 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.1 |
1.2 |
GO:0060134 |
prepulse inhibition(GO:0060134) |
| 0.1 |
0.2 |
GO:0006285 |
base-excision repair, AP site formation(GO:0006285) |
| 0.1 |
1.0 |
GO:0060314 |
regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 |
0.3 |
GO:0051012 |
microtubule sliding(GO:0051012) negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.1 |
0.3 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.1 |
0.8 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 |
0.4 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.1 |
0.2 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.1 |
0.2 |
GO:0034476 |
U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 |
0.5 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.4 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.0 |
0.8 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.5 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.5 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 |
0.9 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.5 |
GO:0051901 |
positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 |
0.4 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.0 |
0.1 |
GO:1904431 |
positive regulation of t-circle formation(GO:1904431) |
| 0.0 |
0.1 |
GO:0002554 |
serotonin secretion by platelet(GO:0002554) |
| 0.0 |
0.1 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 |
0.1 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 |
0.1 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 |
0.2 |
GO:0090309 |
positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 |
0.2 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 |
0.2 |
GO:1900272 |
negative regulation of chronic inflammatory response(GO:0002677) negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.0 |
0.3 |
GO:0043383 |
negative T cell selection(GO:0043383) negative thymic T cell selection(GO:0045060) |
| 0.0 |
0.2 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.0 |
0.1 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.0 |
0.3 |
GO:0090043 |
regulation of tubulin deacetylation(GO:0090043) |
| 0.0 |
0.3 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 |
0.8 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.0 |
0.2 |
GO:0006627 |
protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 |
0.3 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.0 |
0.3 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.1 |
GO:0050689 |
negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 |
0.3 |
GO:0031424 |
keratinization(GO:0031424) |
| 0.0 |
2.3 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.0 |
0.1 |
GO:0001907 |
killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 |
0.3 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.0 |
0.5 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.0 |
0.1 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 |
0.2 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.0 |
0.1 |
GO:0032815 |
negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 |
0.1 |
GO:0071494 |
cellular response to UV-C(GO:0071494) |
| 0.0 |
0.6 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
0.1 |
GO:0032627 |
interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) |
| 0.0 |
0.3 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.0 |
0.3 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.1 |
GO:0006561 |
proline biosynthetic process(GO:0006561) |
| 0.0 |
0.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.2 |
GO:0050650 |
chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 |
0.8 |
GO:0035249 |
synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 |
0.1 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.0 |
0.0 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 |
0.1 |
GO:0031145 |
anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |