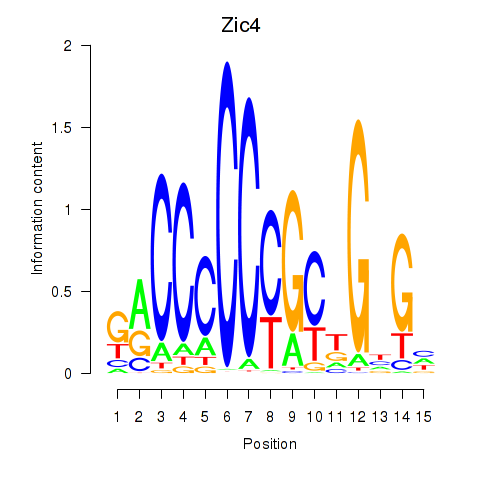
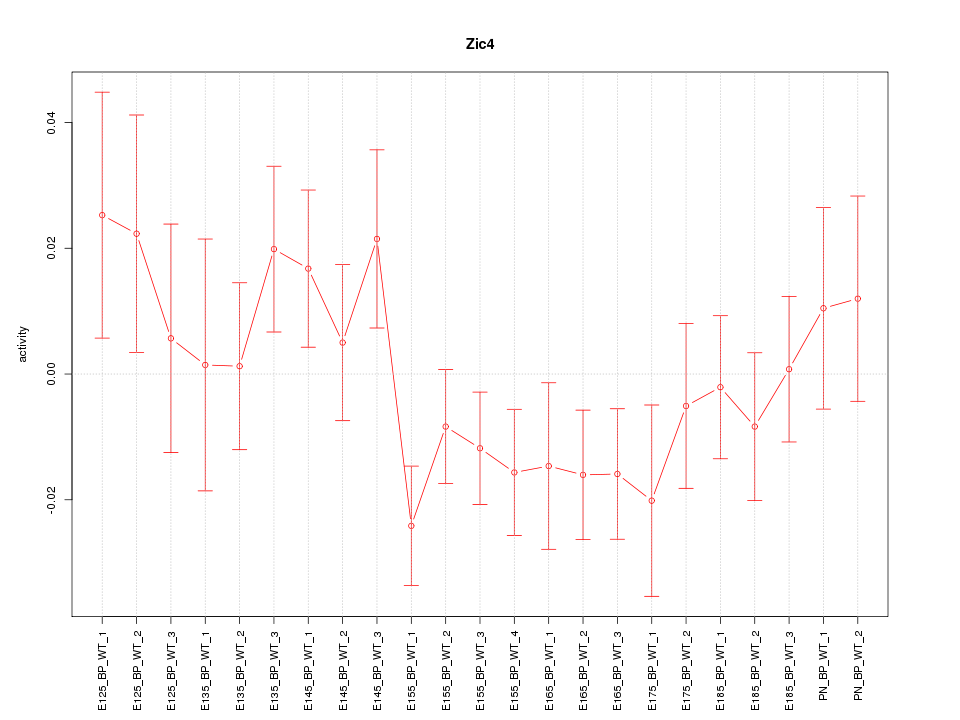
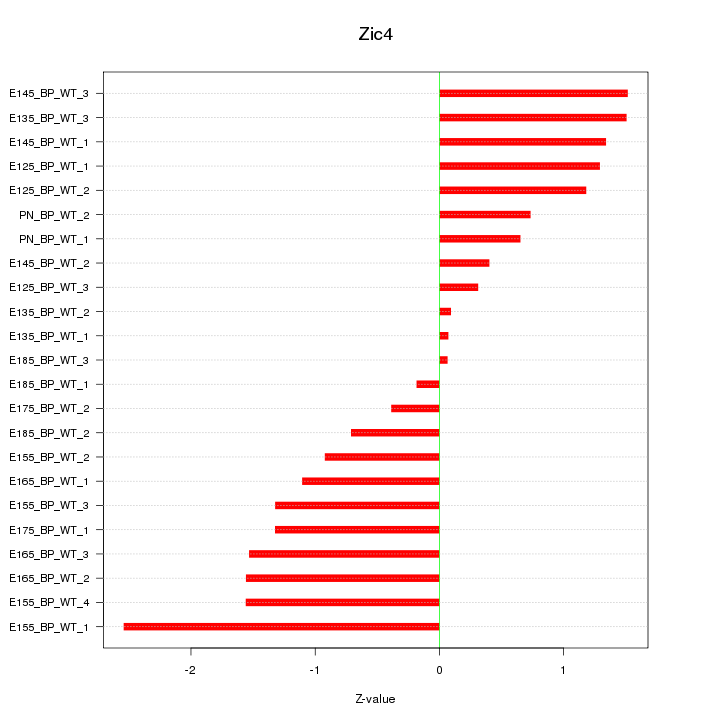
| 0.9 |
7.5 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.6 |
2.6 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.6 |
3.2 |
GO:2001025 |
positive regulation of response to drug(GO:2001025) |
| 0.5 |
2.7 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.5 |
2.1 |
GO:1903802 |
positive regulation of arachidonic acid secretion(GO:0090238) L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.5 |
6.4 |
GO:0060159 |
regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.4 |
2.1 |
GO:0035093 |
spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.4 |
3.2 |
GO:0048170 |
positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.3 |
1.0 |
GO:0097491 |
trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 |
3.8 |
GO:0048484 |
enteric nervous system development(GO:0048484) |
| 0.3 |
1.2 |
GO:0097374 |
sensory neuron axon guidance(GO:0097374) |
| 0.3 |
1.8 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.3 |
0.9 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 0.3 |
1.4 |
GO:0060838 |
lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.2 |
0.9 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 0.2 |
1.2 |
GO:0003199 |
endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 |
0.7 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
0.4 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.2 |
1.2 |
GO:0006452 |
translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.2 |
0.6 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.2 |
0.6 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 0.2 |
0.4 |
GO:0061317 |
canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of heart induction(GO:0090381) |
| 0.2 |
0.9 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.2 |
0.5 |
GO:0002159 |
desmosome assembly(GO:0002159) adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) maintenance of organ identity(GO:0048496) |
| 0.1 |
0.7 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
0.8 |
GO:0002317 |
plasma cell differentiation(GO:0002317) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 |
0.4 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.1 |
1.2 |
GO:0031424 |
keratinization(GO:0031424) establishment of skin barrier(GO:0061436) |
| 0.1 |
0.4 |
GO:0060743 |
epithelial cell maturation involved in prostate gland development(GO:0060743) alveolar secondary septum development(GO:0061144) |
| 0.1 |
0.4 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 |
1.0 |
GO:0097531 |
mast cell migration(GO:0097531) |
| 0.1 |
0.4 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
2.5 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.1 |
2.3 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.1 |
0.8 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.1 |
2.6 |
GO:2000811 |
negative regulation of anoikis(GO:2000811) |
| 0.1 |
0.7 |
GO:0070458 |
detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 |
1.7 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.1 |
1.8 |
GO:0060065 |
uterus development(GO:0060065) |
| 0.1 |
0.4 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 0.1 |
1.7 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
0.2 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.1 |
0.2 |
GO:0009838 |
abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 |
0.1 |
GO:2000173 |
negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.1 |
0.3 |
GO:0010700 |
negative regulation of norepinephrine secretion(GO:0010700) positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 |
0.6 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 |
0.3 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 |
0.3 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.0 |
0.3 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.0 |
1.0 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
1.5 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
0.2 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.7 |
GO:0016188 |
synaptic vesicle maturation(GO:0016188) |
| 0.0 |
0.6 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.0 |
0.2 |
GO:1903377 |
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 |
0.2 |
GO:0070125 |
mitochondrial translational elongation(GO:0070125) |
| 0.0 |
0.3 |
GO:0007042 |
lysosomal lumen acidification(GO:0007042) |
| 0.0 |
5.0 |
GO:0007179 |
transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 |
1.0 |
GO:0042462 |
eye photoreceptor cell development(GO:0042462) |
| 0.0 |
0.2 |
GO:0003161 |
cardiac conduction system development(GO:0003161) |
| 0.0 |
0.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 |
0.3 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.0 |
0.1 |
GO:0015920 |
regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.0 |
0.3 |
GO:0006337 |
nucleosome disassembly(GO:0006337) protein-DNA complex disassembly(GO:0032986) |
| 0.0 |
0.3 |
GO:0035646 |
endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.0 |
0.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.0 |
1.1 |
GO:0006406 |
mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 |
0.5 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.4 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.0 |
0.5 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 |
0.4 |
GO:0035115 |
embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 |
1.1 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
0.5 |
GO:0006301 |
postreplication repair(GO:0006301) |
| 0.0 |
0.7 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 |
0.1 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.0 |
0.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.1 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |