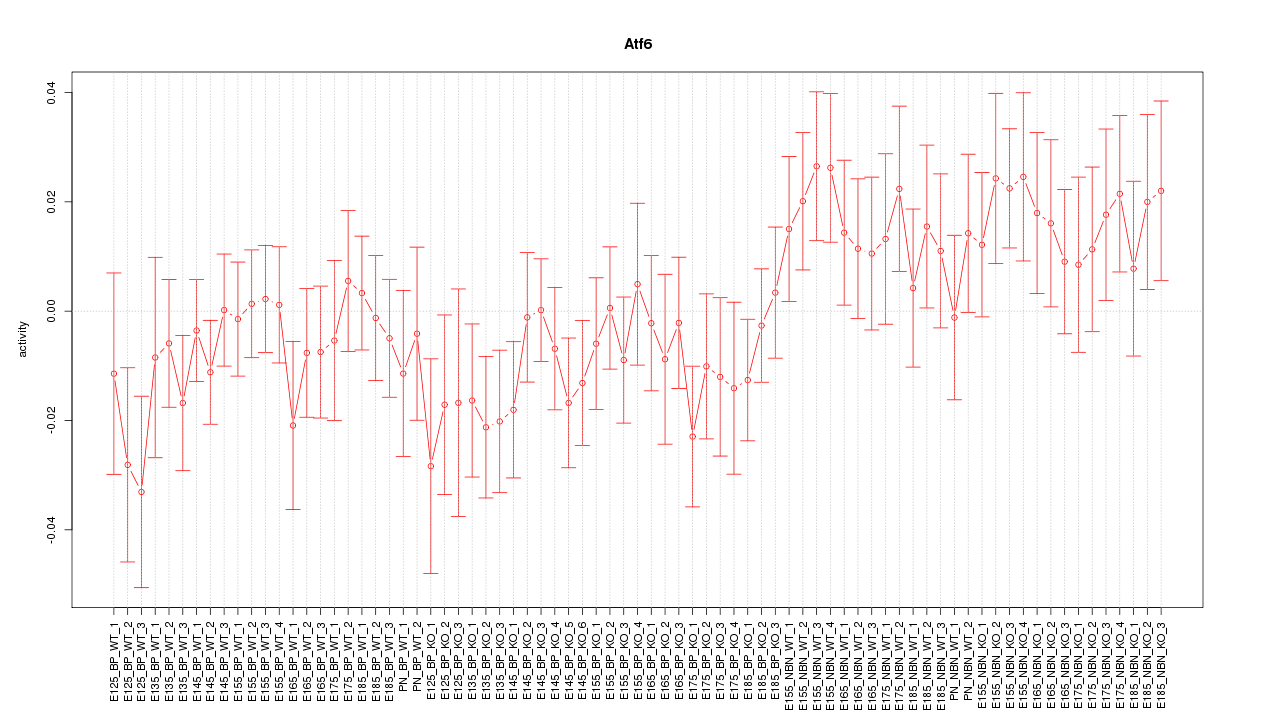
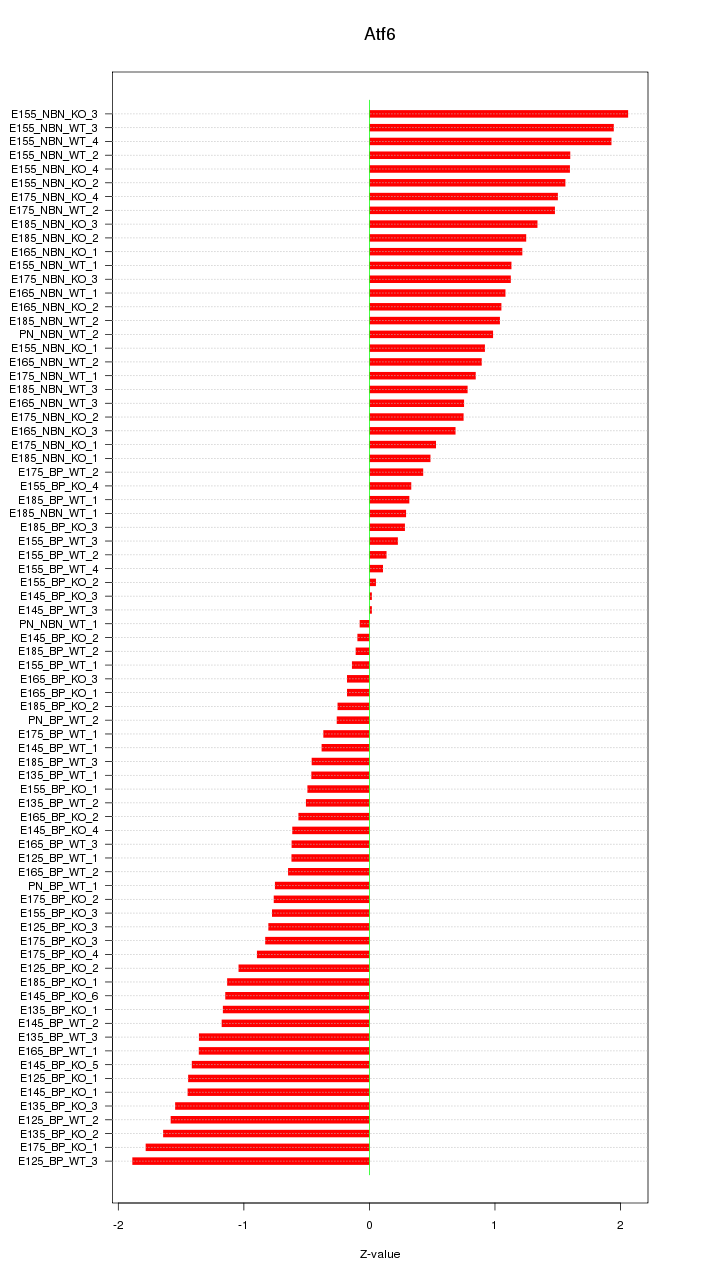
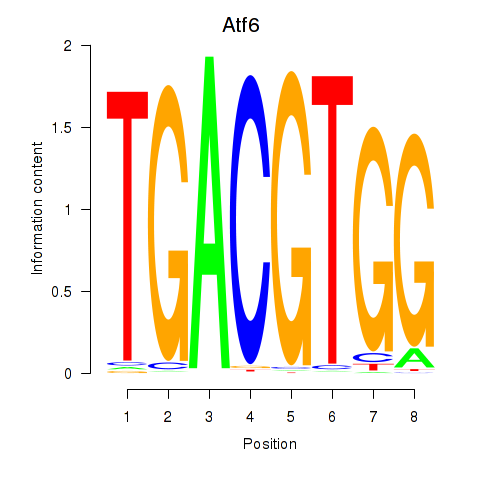
Motif ID: Atf6
Z-value: 1.013
Transcription factors associated with Atf6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Atf6 | ENSMUSG00000026663.6 | Atf6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf6 | mm10_v2_chr1_-_170867761_170867784 | -0.40 | 2.6e-04 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.7 | 23.1 | GO:0043379 | memory T cell differentiation(GO:0043379) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 2.9 | 8.8 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 2.8 | 11.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 2.7 | 13.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 2.4 | 7.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 2.3 | 6.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 2.0 | 11.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 1.9 | 11.4 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 1.7 | 5.0 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 1.6 | 4.7 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.4 | 8.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.3 | 21.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 1.3 | 2.6 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 1.1 | 8.8 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 1.1 | 6.6 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 1.1 | 4.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 1.0 | 4.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.9 | 2.8 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.9 | 2.7 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.8 | 2.5 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.8 | 2.4 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.8 | 38.3 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.8 | 6.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.8 | 2.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.7 | 3.0 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.7 | 2.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.7 | 10.8 | GO:0010881 | regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.7 | 3.6 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.7 | 2.9 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.7 | 2.6 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.7 | 3.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.7 | 0.7 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.6 | 5.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.6 | 8.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.6 | 1.8 | GO:2000331 | regulation of terminal button organization(GO:2000331) |
| 0.6 | 2.9 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.6 | 4.4 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.5 | 5.5 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.5 | 4.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.5 | 1.4 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) |
| 0.5 | 5.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 1.8 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.4 | 1.3 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.4 | 3.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.4 | 2.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 1.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.4 | 2.8 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.4 | 4.0 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.4 | 6.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.4 | 3.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.4 | 10.4 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.4 | 5.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.4 | 1.8 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.4 | 3.5 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.3 | 1.0 | GO:1903862 | flavin-containing compound metabolic process(GO:0042726) positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.3 | 1.7 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.3 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 2.3 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.3 | 1.2 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.3 | 0.9 | GO:2001013 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.3 | 1.4 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.3 | 1.7 | GO:0032811 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.3 | 5.7 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.3 | 5.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.3 | 3.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.2 | 1.0 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.2 | 2.0 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.2 | 1.5 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.2 | 4.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 3.8 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 1.2 | GO:0048133 | NK T cell differentiation(GO:0001865) germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 2.9 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.2 | 2.7 | GO:0043084 | penile erection(GO:0043084) |
| 0.2 | 1.6 | GO:0098712 | L-glutamate import across plasma membrane(GO:0098712) |
| 0.2 | 2.6 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.2 | 1.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 7.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.2 | 0.7 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.2 | 1.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.2 | 7.8 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.2 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 4.4 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.2 | 8.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.2 | 2.3 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.2 | 2.1 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 4.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.8 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 1.0 | GO:0021886 | female meiosis I(GO:0007144) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 1.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 0.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.5 | GO:0032763 | regulation of mast cell cytokine production(GO:0032763) |
| 0.1 | 2.0 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 0.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.4 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 | 1.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) |
| 0.1 | 0.5 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.1 | 2.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.6 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.1 | 1.8 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 5.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.1 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 0.1 | 0.9 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.1 | 1.0 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 2.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 0.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 2.3 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 1.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.3 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 1.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 2.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 0.4 | GO:0046479 | glycolipid catabolic process(GO:0019377) glycosylceramide catabolic process(GO:0046477) glycosphingolipid catabolic process(GO:0046479) |
| 0.1 | 0.5 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 1.7 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 2.6 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.1 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 9.8 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 0.9 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.4 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 0.8 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.1 | 1.1 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 3.4 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.1 | 26.0 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.1 | 2.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.6 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.1 | 0.9 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.4 | GO:0030578 | PML body organization(GO:0030578) |
| 0.1 | 4.5 | GO:0031929 | TOR signaling(GO:0031929) |
| 0.1 | 0.6 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 1.1 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 1.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 1.1 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.0 | 1.5 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 1.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.6 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 1.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.6 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 2.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.6 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 1.0 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 2.9 | GO:0016051 | carbohydrate biosynthetic process(GO:0016051) |
| 0.0 | 0.3 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 2.3 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.6 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 1.8 | 26.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 1.3 | 9.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.8 | 4.0 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.8 | 3.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.7 | 3.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.6 | 9.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.6 | 6.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.5 | 22.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.5 | 2.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 20.9 | GO:0043034 | costamere(GO:0043034) |
| 0.4 | 8.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.4 | 1.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 5.7 | GO:0044754 | autolysosome(GO:0044754) |
| 0.3 | 1.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 1.7 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.3 | 4.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 5.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 0.6 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.3 | 22.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.3 | 7.1 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 0.8 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 0.7 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.2 | 11.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.1 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 0.9 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.2 | 2.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 13.6 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 2.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 6.4 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 8.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 5.0 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.2 | 1.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 2.7 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 9.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 4.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 27.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 5.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.9 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 2.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 4.3 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.1 | 1.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.1 | 5.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 4.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 3.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 3.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 2.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 3.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.1 | GO:0071920 | cleavage body(GO:0071920) |
| 0.0 | 6.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 2.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 5.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 6.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.0 | GO:0018444 | translation release factor complex(GO:0018444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 11.9 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 2.3 | 11.5 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 2.3 | 6.9 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 1.9 | 23.1 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 1.7 | 8.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 1.5 | 6.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.5 | 6.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 1.4 | 5.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.4 | 11.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 1.3 | 6.6 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 1.1 | 4.4 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 1.1 | 3.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 1.0 | 11.4 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 1.0 | 2.9 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.9 | 9.8 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.9 | 6.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.8 | 10.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.8 | 2.4 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.8 | 5.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 2.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 5.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.7 | 2.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.6 | 10.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 1.8 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.6 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.6 | 2.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.5 | 5.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 4.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 5.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.4 | 6.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.4 | 2.7 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 4.8 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 8.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 1.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.3 | 1.7 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 1.7 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 0.8 | GO:1903135 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) cupric ion binding(GO:1903135) |
| 0.3 | 1.0 | GO:0036435 | deubiquitinase activator activity(GO:0035800) K48-linked polyubiquitin binding(GO:0036435) |
| 0.3 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 2.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.2 | 3.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 1.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 2.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.9 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.2 | 10.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 4.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 7.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 18.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.2 | 3.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 2.0 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 1.3 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) neuroligin family protein binding(GO:0097109) |
| 0.2 | 2.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.2 | 2.6 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 0.8 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 3.0 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 2.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.8 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 1.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 6.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.0 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 4.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.8 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.4 | GO:0044729 | double-stranded methylated DNA binding(GO:0010385) hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 1.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.7 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 2.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 2.0 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 2.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.6 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 1.9 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.9 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 1.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0019531 | secondary active sulfate transmembrane transporter activity(GO:0008271) oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.3 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.1 | 1.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.1 | 1.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 17.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 5.3 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 0.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.5 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 6.9 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 1.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.8 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 1.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 10.6 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 6.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 1.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.7 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 25.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.4 | 6.5 | PID_TCR_RAS_PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.3 | 5.0 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.2 | 10.7 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.2 | 11.3 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 5.3 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 7.0 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.1 | PID_IL2_1PATHWAY | IL2-mediated signaling events |
| 0.1 | 2.3 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 2.7 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.1 | 2.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 11.5 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 1.8 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.2 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.7 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 1.4 | PID_NEPHRIN_NEPH1_PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 2.4 | PID_RETINOIC_ACID_PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.9 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.5 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.2 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | ST_DIFFERENTIATION_PATHWAY_IN_PC12_CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.7 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 5.5 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 1.2 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 11.4 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.9 | 6.9 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.8 | 6.4 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.7 | 18.0 | REACTOME_UNBLOCKING_OF_NMDA_RECEPTOR_GLUTAMATE_BINDING_AND_ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.6 | 5.5 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 10.4 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.5 | 6.2 | REACTOME_FGFR1_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.5 | 11.7 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.4 | 2.8 | REACTOME_CYTOSOLIC_SULFONATION_OF_SMALL_MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 2.3 | REACTOME_ORGANIC_CATION_ANION_ZWITTERION_TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.4 | 6.0 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.4 | 5.1 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.3 | 10.3 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 12.9 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.3 | 7.3 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.3 | 10.8 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 14.5 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.2 | 3.4 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 1.8 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 2.6 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 14.1 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 1.5 | REACTOME_NEF_MEDIATED_DOWNREGULATION_OF_MHC_CLASS_I_COMPLEX_CELL_SURFACE_EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 4.7 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 3.1 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.4 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 2.2 | REACTOME_LYSOSOME_VESICLE_BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.8 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.6 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 10.5 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.6 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 2.3 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.6 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 3.7 | REACTOME_LOSS_OF_NLP_FROM_MITOTIC_CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.5 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 2.0 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.8 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 2.3 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION_IN_THE_MEDIAL_TRANS_GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 1.4 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.9 | REACTOME_SYNTHESIS_OF_GLYCOSYLPHOSPHATIDYLINOSITOL_GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME_GROWTH_HORMONE_RECEPTOR_SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 2.6 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.5 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.9 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME_EXTENSION_OF_TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME_SPHINGOLIPID_METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.6 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME_INTERFERON_ALPHA_BETA_SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |