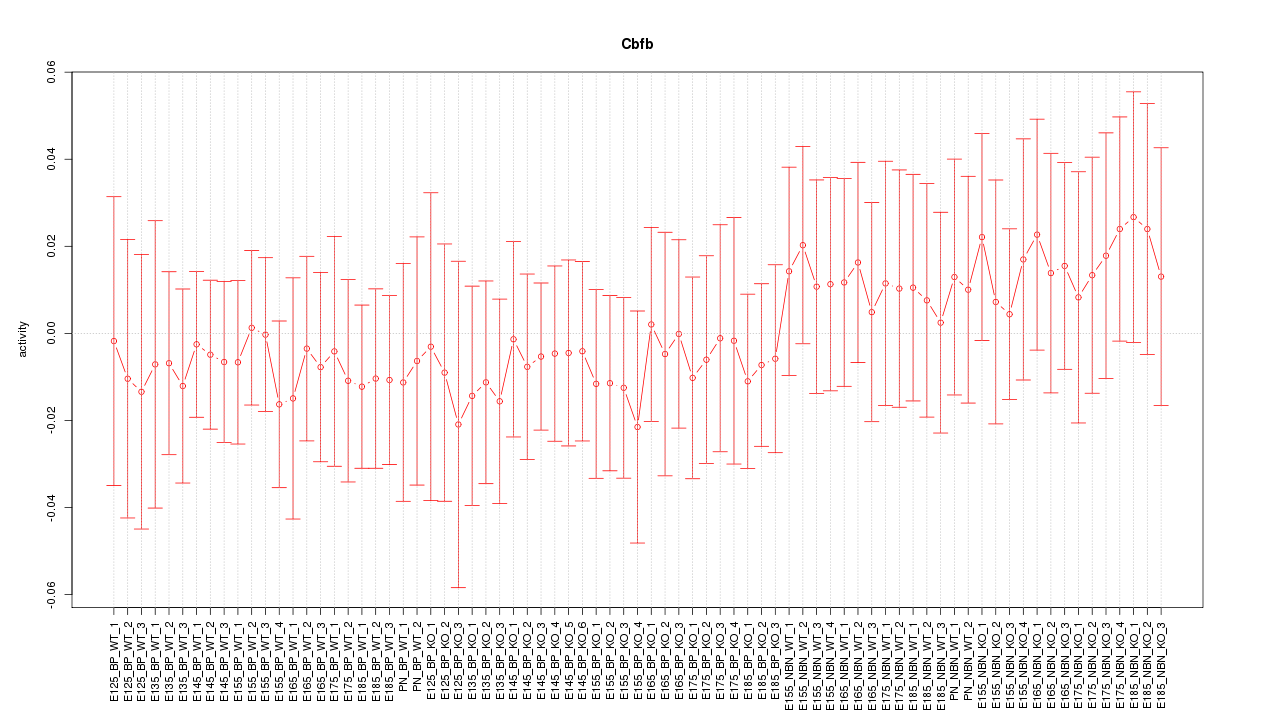
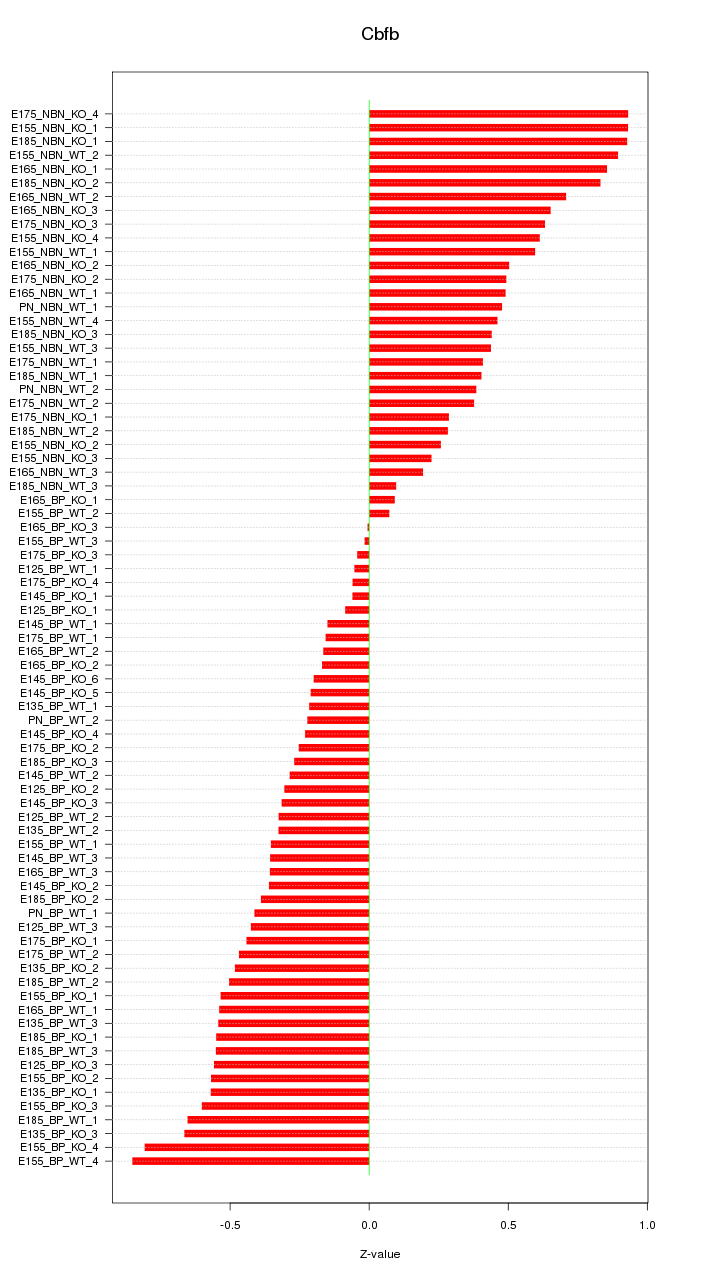
Motif ID: Cbfb
Z-value: 0.474
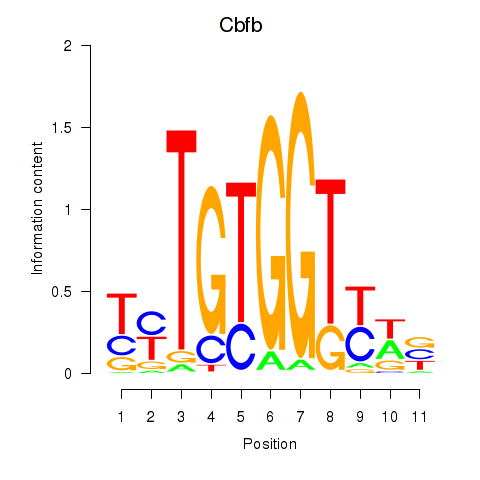
Transcription factors associated with Cbfb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cbfb | ENSMUSG00000031885.7 | Cbfb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cbfb | mm10_v2_chr8_+_105170668_105170686 | -0.27 | 1.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.7 | 2.7 | GO:0035937 | androgen catabolic process(GO:0006710) estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.4 | 3.9 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 1.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 1.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.3 | 2.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.3 | 1.5 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 | 1.9 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.2 | 13.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 0.7 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.1 | 1.2 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.5 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 1.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.6 | GO:0045471 | response to ethanol(GO:0045471) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 13.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 7.9 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 5.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.8 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 11.2 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 14.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.4 | 1.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.2 | 1.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 3.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.2 | 6.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.2 | 7.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.2 | 1.9 | GO:0036122 | BMP binding(GO:0036122) |
| 0.2 | 2.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.7 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.6 | PID_AVB3_OPN_PATHWAY | Osteopontin-mediated events |
| 0.1 | 6.6 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.1 | 13.4 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.1 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.1 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.1 | PID_VEGFR1_PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.6 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 2.1 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 1.9 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 1.9 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.7 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.1 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.2 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 2.7 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME_SYNTHESIS_OF_PA | Genes involved in Synthesis of PA |