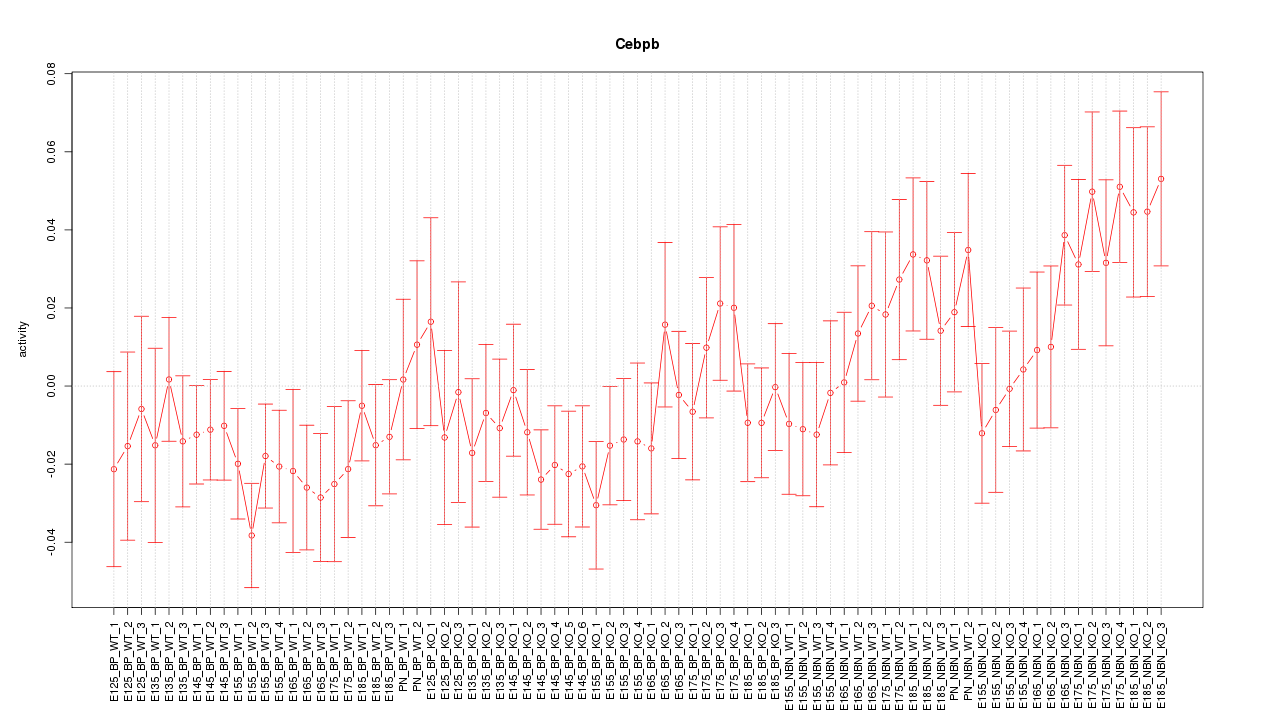
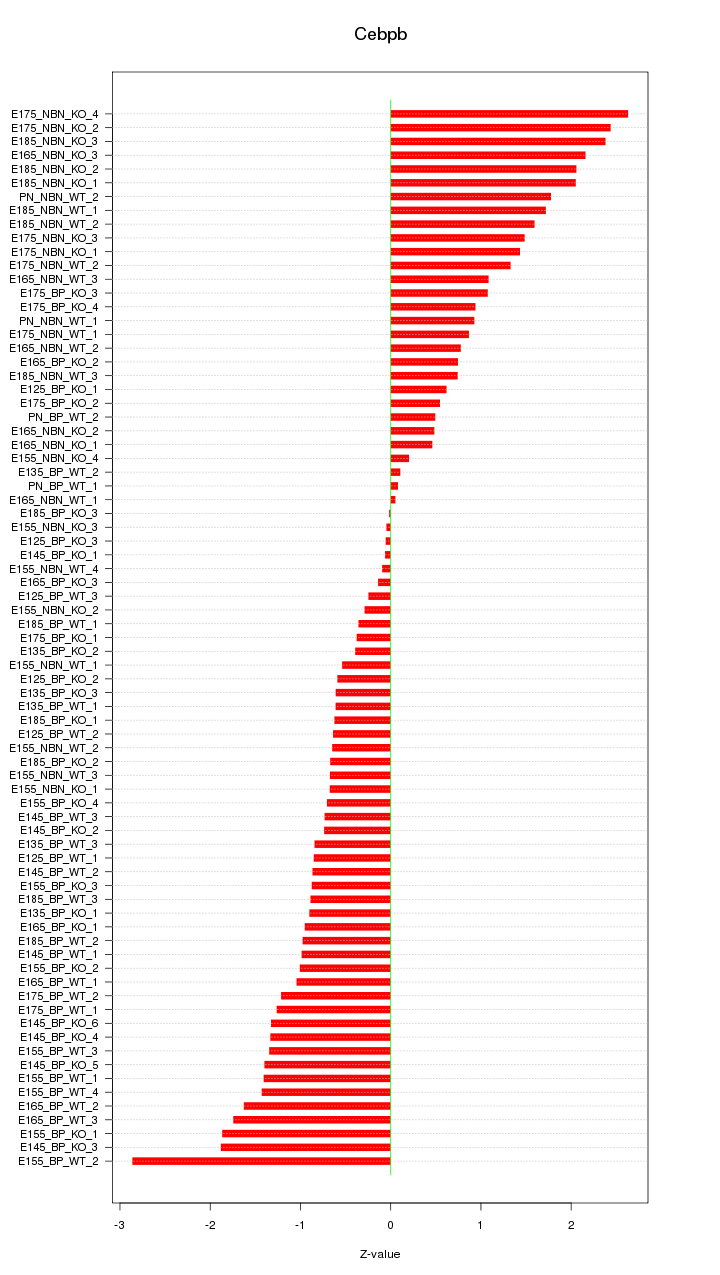
Motif ID: Cebpb
Z-value: 1.169
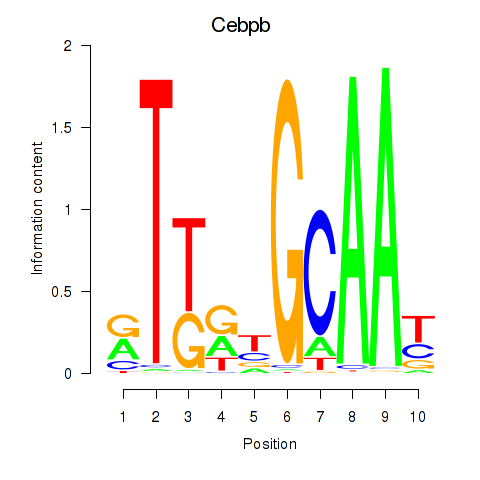
Transcription factors associated with Cebpb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cebpb | ENSMUSG00000056501.3 | Cebpb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cebpb | mm10_v2_chr2_+_167688915_167688973 | 0.74 | 1.4e-14 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 26.3 | GO:0002586 | positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 3.8 | 22.7 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 2.9 | 8.8 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) transforming growth factor-beta secretion(GO:0038044) |
| 2.4 | 7.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 2.4 | 9.5 | GO:1904706 | heme oxidation(GO:0006788) negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 2.3 | 6.8 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 2.1 | 6.3 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 2.1 | 8.2 | GO:0071220 | response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 2.1 | 8.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 1.9 | 9.4 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 1.4 | 7.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 1.4 | 5.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.4 | 4.3 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 1.4 | 4.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 1.4 | 9.9 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 1.3 | 5.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 1.2 | 10.7 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 1.0 | 7.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.0 | 4.9 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 1.0 | 2.9 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.9 | 0.9 | GO:0002580 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) |
| 0.9 | 2.8 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.9 | 3.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 6.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.9 | 6.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.8 | 3.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.8 | 7.3 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.8 | 5.6 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.7 | 2.8 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.7 | 5.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.6 | 1.9 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.6 | 2.5 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.6 | 1.7 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.6 | 2.8 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.6 | 4.5 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.5 | 1.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 4.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 3.5 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.5 | 1.9 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.5 | 7.0 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.4 | 1.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 3.6 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.4 | 4.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 3.7 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 1.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.4 | 3.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 2.0 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.4 | 0.4 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.3 | 1.0 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 | 1.8 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 4.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.3 | 1.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 2.5 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.3 | 2.6 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 10.1 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.2 | 18.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.2 | 5.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.2 | 1.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.2 | 7.3 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.2 | 1.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 1.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.2 | 1.8 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 13.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 1.5 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.2 | 0.6 | GO:0046689 | response to mercury ion(GO:0046689) |
| 0.1 | 0.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 0.5 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.1 | 2.1 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 2.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 2.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 3.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 2.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.3 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.1 | 1.4 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 8.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.5 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 6.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.6 | GO:0007143 | female meiotic division(GO:0007143) |
| 0.1 | 2.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 5.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 1.3 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.1 | 0.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 1.2 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 6.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 8.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.0 | 1.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 2.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.9 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.3 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.6 | GO:0007416 | synapse assembly(GO:0007416) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 9.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 3.2 | 22.7 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 3.1 | 9.4 | GO:0043512 | inhibin A complex(GO:0043512) |
| 2.4 | 7.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 1.4 | 8.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.2 | 6.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 1.2 | 5.8 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.7 | 4.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 4.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.6 | 8.6 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.5 | 1.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.4 | 3.6 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 1.3 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.4 | 1.7 | GO:0071942 | XPC complex(GO:0071942) |
| 0.4 | 7.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.3 | 2.9 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.3 | 4.5 | GO:0043203 | axon hillock(GO:0043203) |
| 0.3 | 1.8 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 7.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 1.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.9 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) tertiary granule(GO:0070820) |
| 0.1 | 11.4 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 8.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 6.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 7.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 10.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 9.5 | GO:0005901 | caveola(GO:0005901) |
| 0.1 | 4.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.9 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.0 | 1.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 3.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 3.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 4.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.1 | GO:0044754 | secondary lysosome(GO:0005767) autolysosome(GO:0044754) |
| 0.0 | 0.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.6 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.6 | GO:0005819 | spindle(GO:0005819) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 35.6 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 2.5 | 9.9 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 2.4 | 7.3 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 2.4 | 9.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 1.8 | 7.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 1.6 | 9.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.5 | 4.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.5 | 8.8 | GO:0045340 | mercury ion binding(GO:0045340) |
| 1.4 | 5.6 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 1.4 | 6.8 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.2 | 4.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 1.1 | 4.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.9 | 2.8 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.9 | 22.7 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.9 | 5.6 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.9 | 2.8 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.9 | 5.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.8 | 1.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.7 | 3.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.6 | 2.5 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.6 | 1.8 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.6 | 18.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.5 | 3.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.5 | 6.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.5 | 1.9 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.5 | 3.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.5 | 3.7 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 8.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.4 | 2.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.4 | 2.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.4 | 10.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 0.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.3 | 4.9 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.3 | 1.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 2.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.3 | 1.9 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.3 | 1.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 2.5 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.2 | 6.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 2.9 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 1.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 4.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 7.3 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.2 | 2.4 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 5.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 11.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 5.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 2.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 5.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 1.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.1 | 11.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 3.0 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 2.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 3.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.4 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 0.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 3.4 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 2.9 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 1.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 8.2 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 10.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 5.8 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.9 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 6.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.3 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.5 | 17.0 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.5 | 8.8 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 9.3 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.3 | 6.8 | PID_PDGFRA_PATHWAY | PDGFR-alpha signaling pathway |
| 0.3 | 9.7 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.3 | 9.4 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.3 | 13.9 | PID_IL12_2PATHWAY | IL12-mediated signaling events |
| 0.2 | 9.9 | PID_SYNDECAN_1_PATHWAY | Syndecan-1-mediated signaling events |
| 0.2 | 9.5 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.2 | 14.6 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 7.9 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.2 | 1.8 | SA_FAS_SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 4.2 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 19.1 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.9 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 6.1 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.8 | PID_CDC42_REG_PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.9 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 2.6 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID_IL6_7_PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.3 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 4.3 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 9.4 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 1.0 | 26.3 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.8 | 9.3 | REACTOME_IKK_COMPLEX_RECRUITMENT_MEDIATED_BY_RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.8 | 14.4 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 5.8 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.5 | 14.7 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.5 | 12.0 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.5 | 7.5 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.5 | 5.2 | REACTOME_P130CAS_LINKAGE_TO_MAPK_SIGNALING_FOR_INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 10.7 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 30.0 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.3 | 11.8 | REACTOME_AMINO_ACID_TRANSPORT_ACROSS_THE_PLASMA_MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.3 | 7.3 | REACTOME_ACTIVATED_AMPK_STIMULATES_FATTY_ACID_OXIDATION_IN_MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.3 | 2.6 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 8.2 | REACTOME_INTEGRIN_ALPHAIIB_BETA3_SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.3 | 9.9 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 6.6 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 16.9 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 17.1 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 4.1 | REACTOME_ACTIVATION_OF_GENES_BY_ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.2 | 6.3 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 1.8 | REACTOME_EXTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 3.0 | REACTOME_METAL_ION_SLC_TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.6 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 4.9 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.2 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.7 | REACTOME_FORMATION_OF_INCISION_COMPLEX_IN_GG_NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 6.8 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.5 | REACTOME_INHIBITION_OF_VOLTAGE_GATED_CA2_CHANNELS_VIA_GBETA_GAMMA_SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.2 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.2 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.0 | REACTOME_TOLL_RECEPTOR_CASCADES | Genes involved in Toll Receptor Cascades |
| 0.0 | 2.9 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME_PHASE1_FUNCTIONALIZATION_OF_COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |