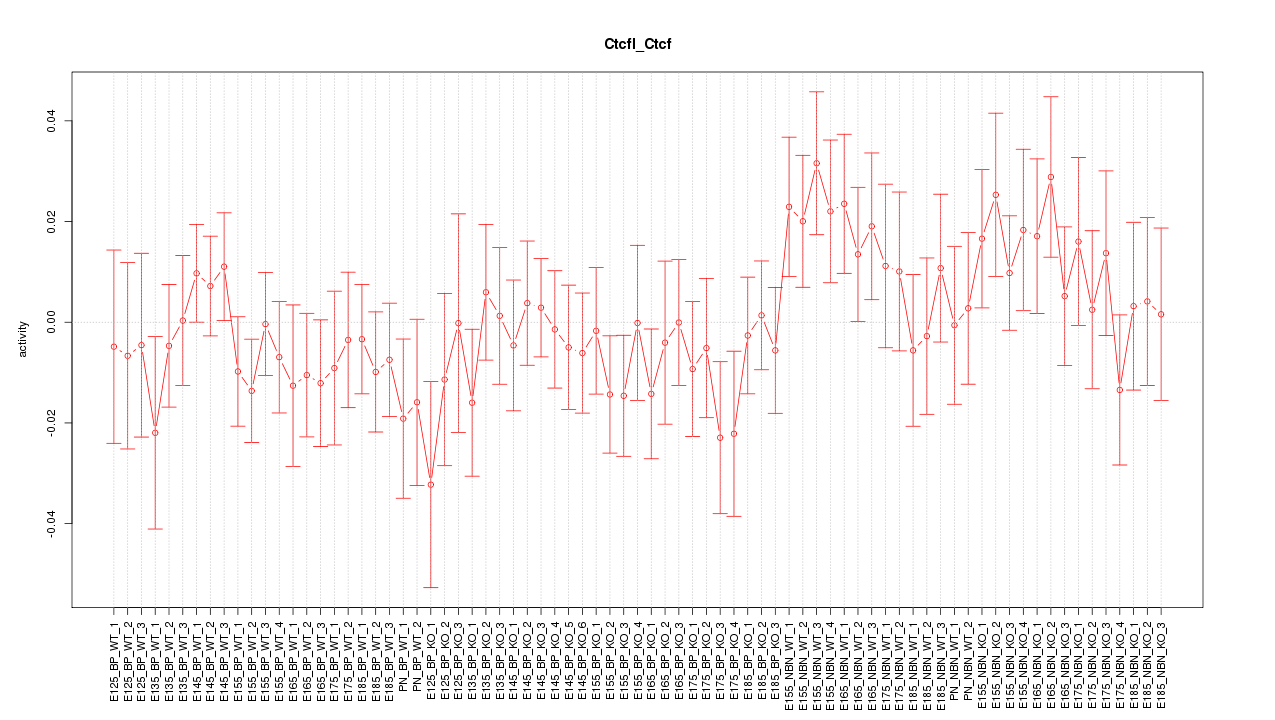
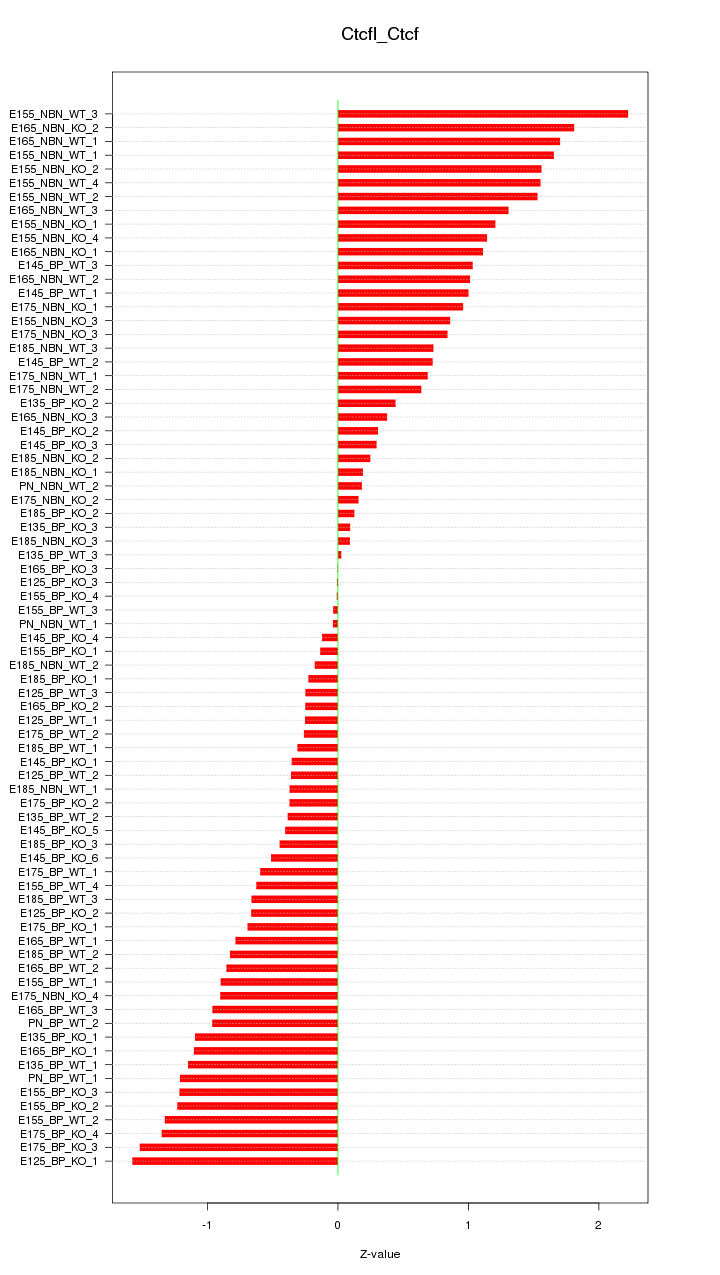
Motif ID: Ctcfl_Ctcf
Z-value: 0.889
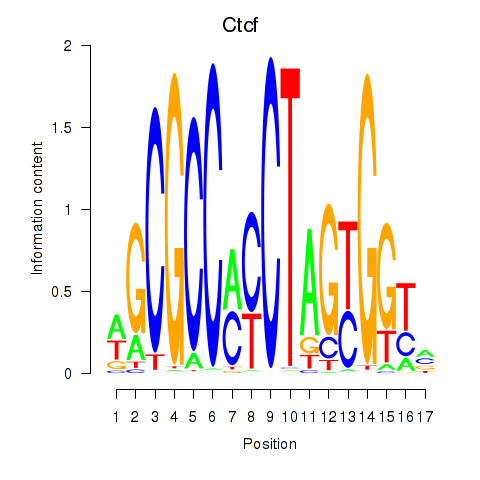
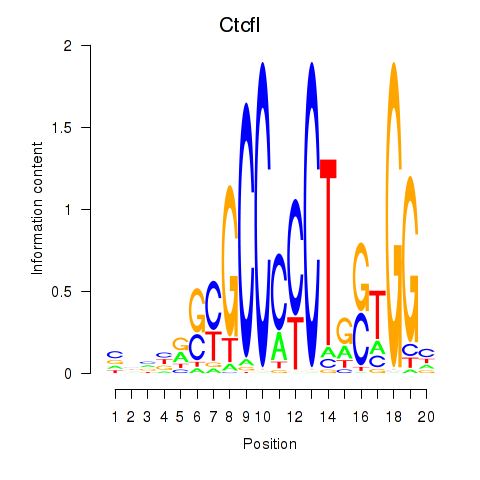
Transcription factors associated with Ctcfl_Ctcf:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ctcf | ENSMUSG00000005698.9 | Ctcf |
| Ctcfl | ENSMUSG00000070495.5 | Ctcfl |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ctcfl | mm10_v2_chr2_-_173119402_173119525 | -0.26 | 2.2e-02 | Click! |
| Ctcf | mm10_v2_chr8_+_105636509_105636589 | -0.23 | 4.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.7 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 3.5 | 10.6 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 3.1 | 12.3 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 3.1 | 12.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 2.7 | 8.2 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 2.7 | 10.8 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 2.6 | 7.9 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 2.3 | 4.7 | GO:0042323 | negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.3 | 16.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 2.1 | 19.1 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 2.1 | 6.3 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 1.7 | 8.7 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 1.6 | 6.5 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 1.6 | 4.9 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 1.5 | 7.7 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 1.5 | 4.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 1.5 | 4.4 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.4 | 5.6 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 1.1 | 6.7 | GO:0032439 | endosome localization(GO:0032439) |
| 1.0 | 3.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.9 | 4.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.8 | 4.1 | GO:0010637 | diet induced thermogenesis(GO:0002024) negative regulation of mitochondrial fusion(GO:0010637) |
| 0.8 | 6.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.8 | 6.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.8 | 2.3 | GO:0045113 | regulation of integrin biosynthetic process(GO:0045113) |
| 0.7 | 2.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.7 | 2.2 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.7 | 2.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.7 | 2.1 | GO:0003289 | endocardial cushion fusion(GO:0003274) atrial septum primum morphogenesis(GO:0003289) |
| 0.7 | 2.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.6 | 4.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.6 | 1.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.5 | 2.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.5 | 1.6 | GO:0009826 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.5 | 4.3 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.5 | 23.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.5 | 5.0 | GO:0030432 | peristalsis(GO:0030432) |
| 0.5 | 1.5 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.5 | 2.8 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.4 | 3.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.3 | GO:0009838 | abscission(GO:0009838) positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.4 | 2.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.4 | 3.9 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.4 | 3.0 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 1.3 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.4 | 2.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.4 | 1.9 | GO:0001705 | ectoderm formation(GO:0001705) ectodermal cell differentiation(GO:0010668) |
| 0.4 | 1.1 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) |
| 0.4 | 2.9 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.4 | 3.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.4 | 1.4 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.3 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.3 | 2.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.3 | 1.4 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.3 | 1.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.3 | 2.0 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.3 | 1.0 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.3 | 1.0 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 0.9 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 1.9 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 0.6 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 0.9 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.3 | 3.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.3 | 1.8 | GO:0097646 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.3 | 1.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.3 | 0.8 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.3 | 4.8 | GO:0042297 | vocal learning(GO:0042297) imitative learning(GO:0098596) |
| 0.3 | 1.6 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.3 | 5.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 1.0 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.2 | 1.9 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.2 | 0.7 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.2 | 0.7 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.2 | 2.9 | GO:0043252 | oligopeptide transport(GO:0006857) prostaglandin transport(GO:0015732) sodium-independent organic anion transport(GO:0043252) |
| 0.2 | 2.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 1.7 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.2 | 0.6 | GO:0019230 | proprioception(GO:0019230) |
| 0.2 | 0.8 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 0.8 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) |
| 0.2 | 0.8 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.2 | 1.6 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 1.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.2 | 1.7 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 | 0.6 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 2.6 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 2.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.2 | 1.9 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.2 | 2.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 0.7 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.2 | 4.3 | GO:0030818 | negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of cAMP biosynthetic process(GO:0030818) |
| 0.2 | 1.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 25.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.2 | 3.5 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 2.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.2 | 3.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 4.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.2 | 2.6 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.1 | 2.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 3.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.9 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.1 | 1.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.6 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 0.4 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.1 | 0.5 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.0 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.1 | 0.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 2.6 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 3.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 1.8 | GO:0036296 | cellular response to increased oxygen levels(GO:0036295) response to increased oxygen levels(GO:0036296) |
| 0.1 | 0.9 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 8.6 | GO:0061178 | regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.3 | GO:0015870 | acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 | 0.4 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.9 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 1.9 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.1 | 1.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 2.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.1 | 2.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.7 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 3.0 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.1 | 0.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.1 | 0.6 | GO:1901299 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) negative regulation of hydrogen peroxide-mediated programmed cell death(GO:1901299) |
| 0.1 | 1.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.6 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.5 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.7 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 0.1 | 5.5 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.1 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.4 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 2.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.0 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 2.0 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.1 | 0.7 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 2.9 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.7 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 1.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.5 | GO:0015813 | L-glutamate transport(GO:0015813) |
| 0.0 | 1.4 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.3 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.5 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.0 | 0.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 4.7 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.6 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) |
| 0.0 | 0.5 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.7 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 1.4 | GO:0006364 | rRNA processing(GO:0006364) rRNA metabolic process(GO:0016072) |
| 0.0 | 1.6 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 1.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0090201 | negative regulation of release of cytochrome c from mitochondria(GO:0090201) |
| 0.0 | 0.9 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.7 | GO:0042755 | eating behavior(GO:0042755) |
| 0.0 | 0.1 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.4 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 3.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 1.5 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 2.3 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.9 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 2.7 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 1.7 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.6 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.7 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) |
| 0.0 | 0.2 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.6 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.4 | GO:0097006 | regulation of plasma lipoprotein particle levels(GO:0097006) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.4 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 19.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 1.5 | 4.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.4 | 22.5 | GO:0043196 | varicosity(GO:0043196) |
| 1.4 | 10.9 | GO:0097433 | dense body(GO:0097433) |
| 1.3 | 6.6 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.1 | 5.5 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 1.0 | 2.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
| 0.9 | 4.3 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.8 | 2.4 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.7 | 10.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 4.9 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.7 | 2.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.6 | 21.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.6 | 3.7 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 3.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.6 | 3.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.6 | 2.9 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 3.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.5 | 2.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.5 | 2.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.5 | 1.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.5 | 9.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.4 | 2.2 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.4 | 2.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 2.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 1.7 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 1.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 0.9 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 0.6 | GO:0031523 | Myb complex(GO:0031523) |
| 0.3 | 4.7 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 1.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.2 | 22.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 0.7 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.2 | 2.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.2 | 3.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.2 | 2.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.8 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 2.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.2 | 1.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.2 | 0.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 1.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 2.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 0.9 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 1.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 5.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.2 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 10.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.3 | GO:0090543 | ESCRT III complex(GO:0000815) Flemming body(GO:0090543) |
| 0.1 | 0.1 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 3.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 1.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.0 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 0.4 | GO:0030677 | ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 5.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 18.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 7.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 1.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 2.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 1.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 1.0 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 1.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 4.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 4.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 11.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 11.4 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 7.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 5.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.7 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 6.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 25.4 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.2 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 9.5 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.0 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 19.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 2.7 | 27.2 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 2.6 | 7.9 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 2.2 | 6.6 | GO:0071209 | U7 snRNA binding(GO:0071209) |
| 2.2 | 6.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 2.1 | 8.2 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 1.9 | 7.5 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.2 | 5.0 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 1.1 | 5.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.0 | 6.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.9 | 4.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 10.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.8 | 3.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.7 | 2.2 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.7 | 2.9 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.7 | 2.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.7 | 2.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.7 | 4.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.6 | 5.4 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.6 | 2.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 0.5 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.5 | 2.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.5 | 4.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.5 | 1.0 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) epinephrine binding(GO:0051379) |
| 0.5 | 1.5 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.5 | 0.9 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.4 | 1.7 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 2.2 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.4 | 7.9 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.4 | 2.9 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.4 | 1.2 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.4 | 10.9 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.4 | 2.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 2.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 1.7 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.3 | 2.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.3 | 1.0 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.3 | 1.0 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.3 | 4.9 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.3 | 2.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 1.0 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 1.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.2 | 1.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 3.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.2 | 1.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.2 | 2.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.2 | 2.2 | GO:0044548 | potassium ion leak channel activity(GO:0022841) S100 protein binding(GO:0044548) |
| 0.2 | 2.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 0.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 0.5 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 0.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 5.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.2 | 0.6 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.2 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 10.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 21.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 5.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.9 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 4.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 3.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.6 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 5.0 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.5 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 13.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 5.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.9 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 1.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 4.3 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.1 | 2.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.7 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 3.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 0.4 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 27.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 0.4 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 1.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 5.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.2 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 1.6 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 2.2 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.1 | 1.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 5.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 3.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.1 | 2.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) flap endonuclease activity(GO:0048256) |
| 0.0 | 3.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 2.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 4.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 3.8 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 1.8 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0004673 | phosphorelay sensor kinase activity(GO:0000155) protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) |
| 0.0 | 0.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 0.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.7 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.6 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.0 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.0 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 21.9 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 1.0 | 19.1 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.6 | 25.1 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.4 | 6.4 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.4 | 12.0 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.2 | 9.1 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.2 | 2.1 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.1 | 2.5 | SA_PROGRAMMED_CELL_DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.3 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 8.7 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.6 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 4.3 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 2.1 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 4.6 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 7.2 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.0 | PID_ANTHRAX_PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.2 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 1.5 | PID_PRL_SIGNALING_EVENTS_PATHWAY | Signaling events mediated by PRL |
| 0.1 | 11.7 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 9.5 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 4.3 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 3.1 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 11.8 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 1.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.5 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 2.7 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.2 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID_CXCR3_PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 17.0 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.8 | 19.1 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 15.1 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 1.0 | REACTOME_EICOSANOID_LIGAND_BINDING_RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.5 | 8.6 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.5 | 12.3 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 10.8 | REACTOME_CREB_PHOSPHORYLATION_THROUGH_THE_ACTIVATION_OF_CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.4 | 2.1 | REACTOME_ANDROGEN_BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.4 | 2.9 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 0.4 | 4.7 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.3 | 10.6 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.3 | 2.2 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 6.4 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 5.7 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.2 | 2.5 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 7.9 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 4.5 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 2.2 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.2 | 6.8 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 6.5 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.2 | 2.3 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 24.6 | REACTOME_G_ALPHA_I_SIGNALLING_EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.2 | REACTOME_CA_DEPENDENT_EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 2.5 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.2 | 1.4 | REACTOME_PHOSPHORYLATION_OF_CD3_AND_TCR_ZETA_CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.9 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.5 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 21.0 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 8.4 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.4 | REACTOME_FRS2_MEDIATED_CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 4.5 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.2 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 5.1 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 2.0 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 2.0 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 1.0 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.3 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 9.0 | REACTOME_SRP_DEPENDENT_COTRANSLATIONAL_PROTEIN_TARGETING_TO_MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 1.2 | REACTOME_N_GLYCAN_ANTENNAE_ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.4 | REACTOME_ENDOSOMAL_SORTING_COMPLEX_REQUIRED_FOR_TRANSPORT_ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 2.2 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.3 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.8 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.0 | REACTOME_PYRUVATE_METABOLISM_AND_CITRIC_ACID_TCA_CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.7 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.6 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.0 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.3 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME_SPHINGOLIPID_DE_NOVO_BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.9 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.8 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.0 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME_MTORC1_MEDIATED_SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.4 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME_SEMA4D_IN_SEMAPHORIN_SIGNALING | Genes involved in Sema4D in semaphorin signaling |