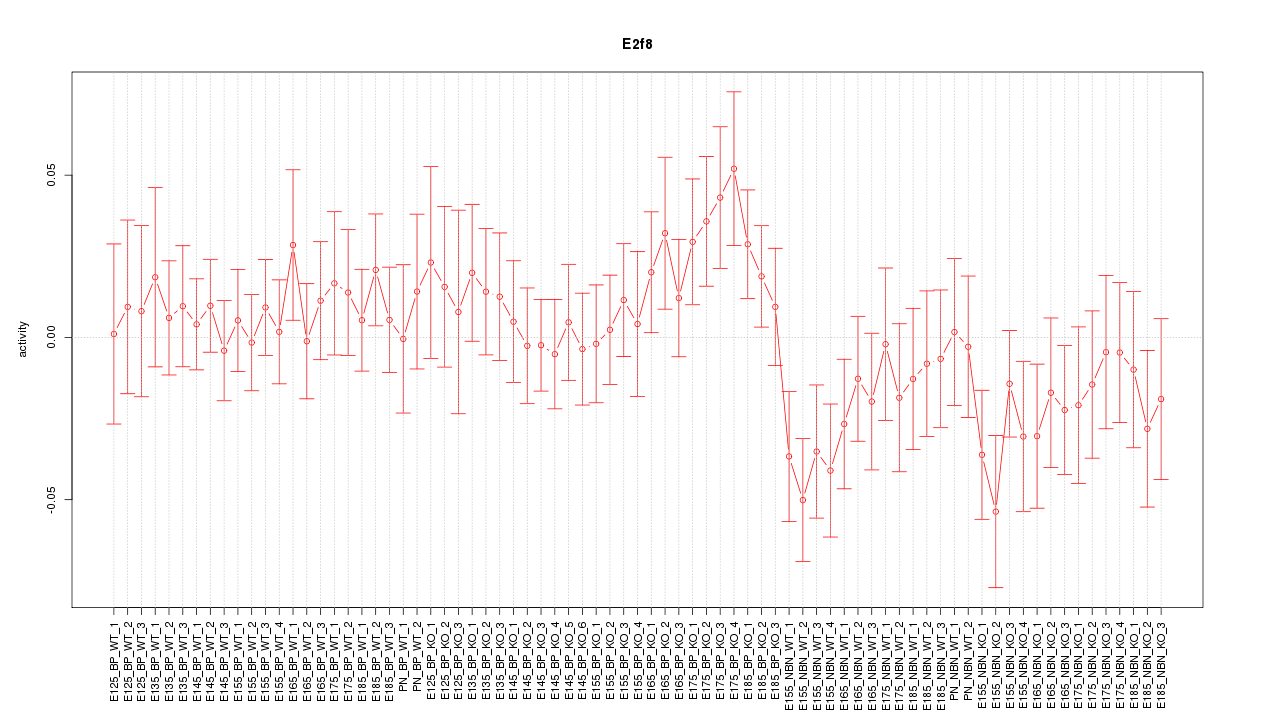
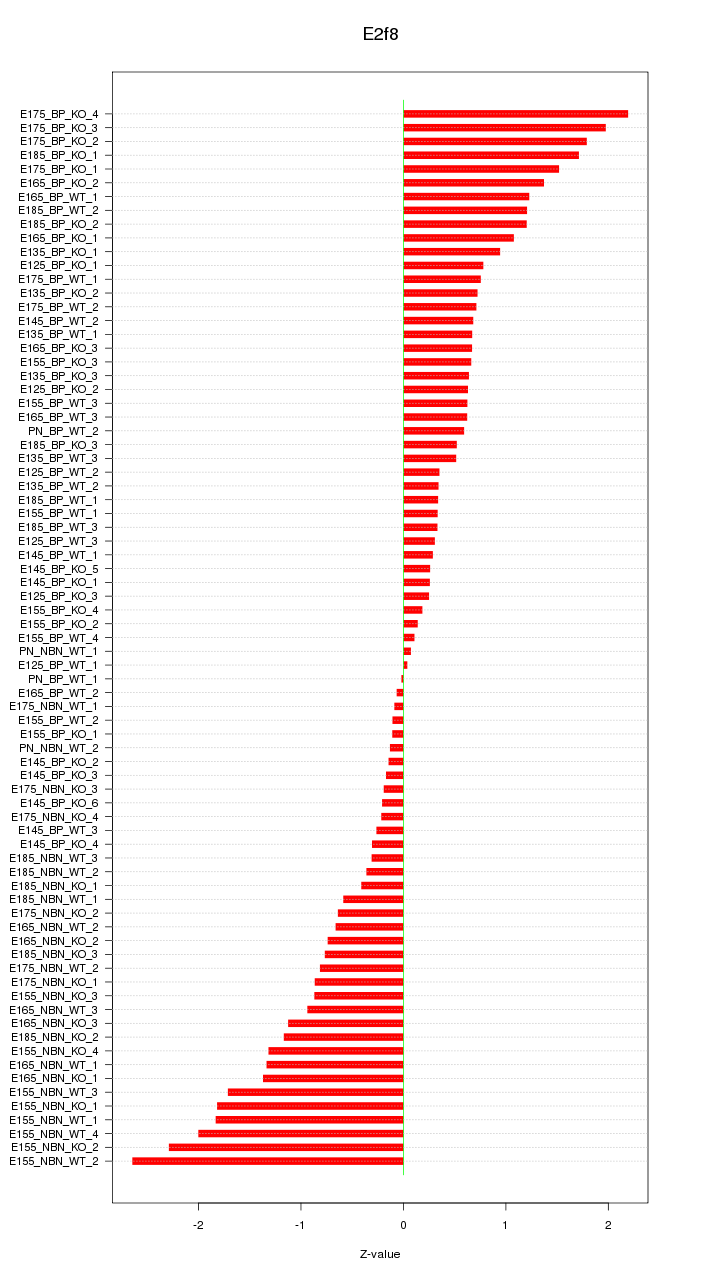
Motif ID: E2f8
Z-value: 0.974
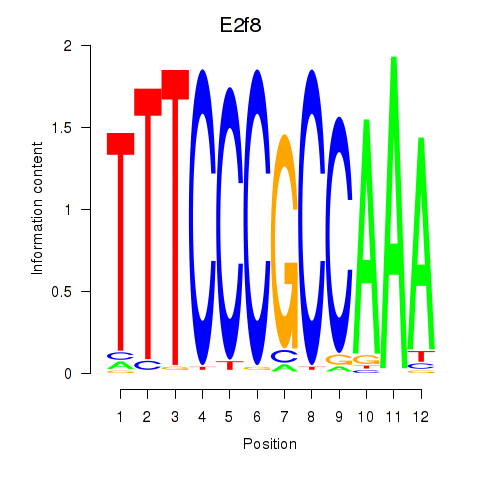
Transcription factors associated with E2f8:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f8 | ENSMUSG00000046179.11 | E2f8 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f8 | mm10_v2_chr7_-_48881596_48881619 | 0.85 | 4.2e-22 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 2.7 | 8.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 2.6 | 7.9 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 2.2 | 33.4 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 2.2 | 10.9 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.6 | 15.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 1.2 | 3.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 1.1 | 3.4 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 1.1 | 12.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.9 | 16.9 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.9 | 9.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.8 | 10.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.7 | 5.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.7 | 2.2 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.7 | 3.6 | GO:0002121 | inter-male aggressive behavior(GO:0002121) response to pheromone(GO:0019236) |
| 0.6 | 3.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) |
| 0.6 | 15.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.5 | 1.6 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 3.6 | GO:0046549 | phenol-containing compound catabolic process(GO:0019336) retinal cone cell development(GO:0046549) |
| 0.5 | 1.5 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.4 | 3.0 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 1.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.3 | 2.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.7 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.2 | 3.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.2 | 1.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.2 | 6.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 1.0 | GO:0002326 | B cell lineage commitment(GO:0002326) ectopic germ cell programmed cell death(GO:0035234) |
| 0.1 | 7.5 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 1.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 16.0 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.1 | 4.4 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.8 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) regulation of ribonucleoprotein complex localization(GO:2000197) |
| 0.1 | 3.5 | GO:0033045 | regulation of sister chromatid segregation(GO:0033045) |
| 0.1 | 0.5 | GO:0002317 | plasma cell differentiation(GO:0002317) positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.5 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 8.2 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.0 | 8.8 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.1 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:1990423 | RZZ complex(GO:1990423) |
| 3.4 | 16.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 2.7 | 8.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 2.1 | 6.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.6 | 7.9 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.5 | 7.5 | GO:0031523 | Myb complex(GO:0031523) |
| 1.4 | 20.7 | GO:0042555 | MCM complex(GO:0042555) |
| 1.1 | 3.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.8 | 2.4 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.6 | 13.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.5 | 36.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.4 | 3.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.4 | 10.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.3 | 1.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.2 | 8.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.0 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 3.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 1.0 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 3.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 1.5 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 0.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 27.3 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 7.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.2 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.6 | 22.5 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 3.4 | 16.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 2.8 | 8.3 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 1.6 | 22.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 1.4 | 11.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 1.0 | 3.0 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.8 | 6.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 3.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.4 | 2.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 3.4 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.3 | 3.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.3 | 1.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.3 | 1.7 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.9 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.5 | GO:0050786 | icosatetraenoic acid binding(GO:0050543) arachidonic acid binding(GO:0050544) RAGE receptor binding(GO:0050786) |
| 0.2 | 11.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 1.0 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.2 | 2.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.2 | 7.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.2 | 4.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 28.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 6.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 3.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 3.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 0.5 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 10.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 3.4 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.0 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 7.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 2.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 2.3 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.6 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.6 | 7.9 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 20.8 | PID_ATR_PATHWAY | ATR signaling pathway |
| 0.2 | 10.9 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.2 | 18.3 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.1 | 2.4 | PID_RANBP2_PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 8.8 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.6 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.5 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.0 | 2.2 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 1.0 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID_FAS_PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID_RAS_PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 41.5 | REACTOME_G1_S_SPECIFIC_TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 2.1 | 18.6 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.5 | 21.6 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.6 | 5.8 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.6 | 10.3 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.4 | 1.6 | REACTOME_PROCESSIVE_SYNTHESIS_ON_THE_LAGGING_STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.3 | 3.6 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 3.0 | REACTOME_SLBP_DEPENDENT_PROCESSING_OF_REPLICATION_DEPENDENT_HISTONE_PRE_MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.6 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 2.4 | REACTOME_MICRORNA_MIRNA_BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.0 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 6.0 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.0 | REACTOME_DOUBLE_STRAND_BREAK_REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 3.6 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.2 | REACTOME_SIGNALING_BY_EGFR_IN_CANCER | Genes involved in Signaling by EGFR in Cancer |