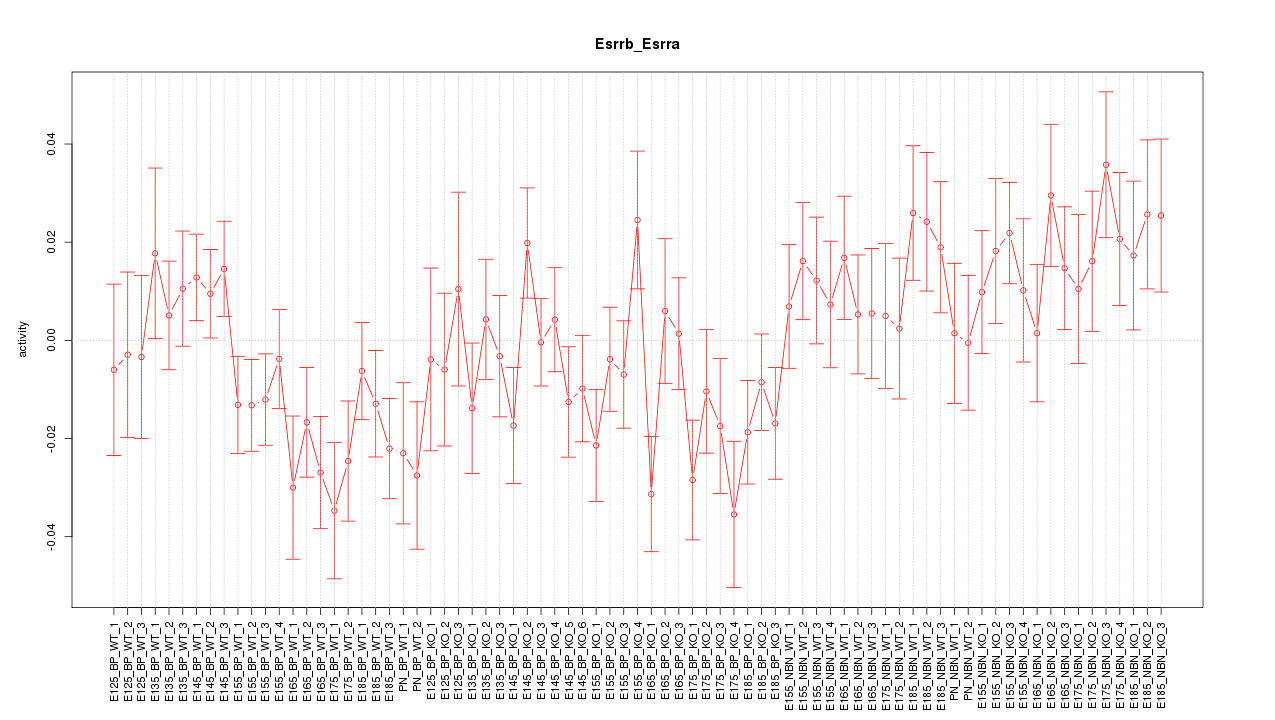
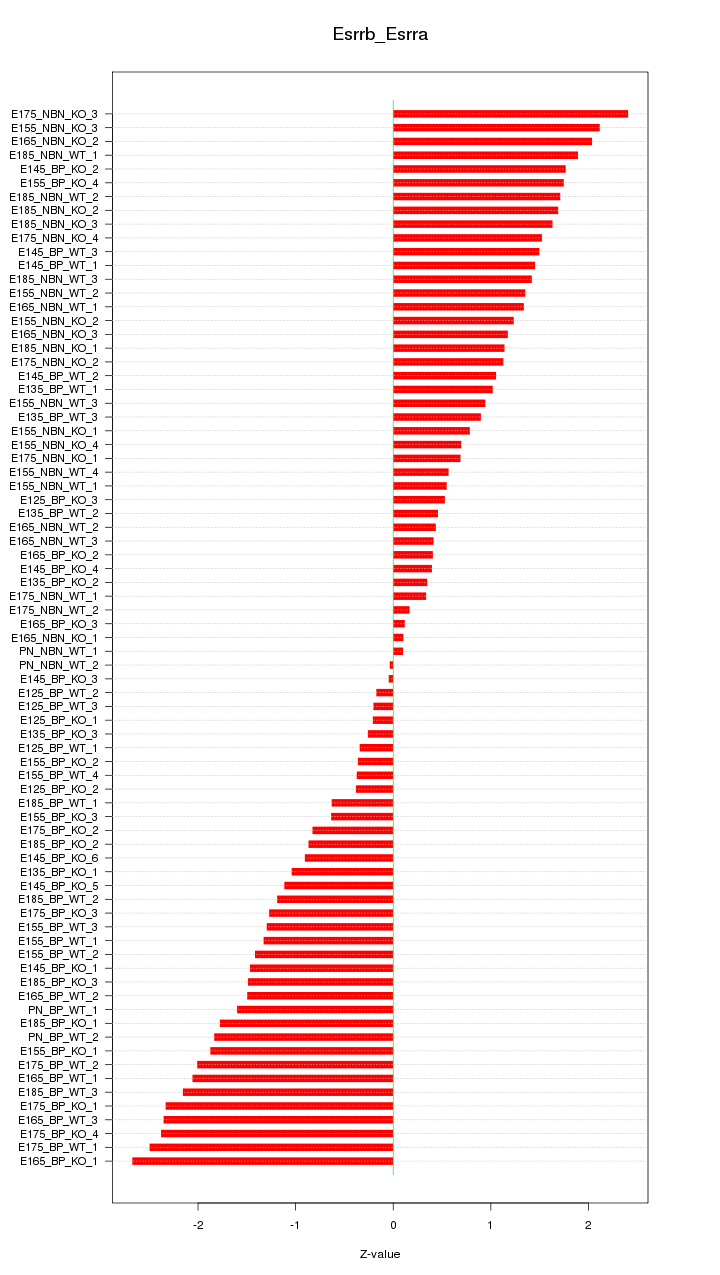
Motif ID: Esrrb_Esrra
Z-value: 1.322
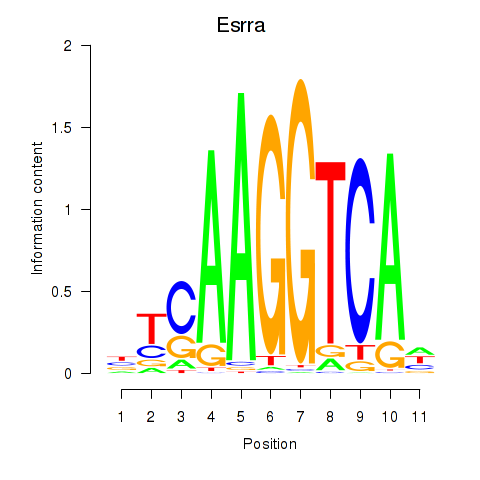
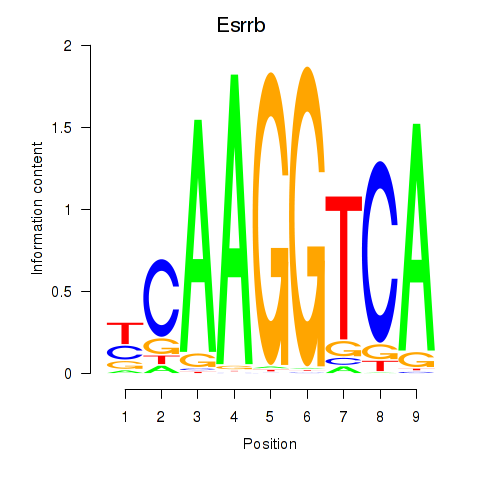
Transcription factors associated with Esrrb_Esrra:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Esrra | ENSMUSG00000024955.7 | Esrra |
| Esrrb | ENSMUSG00000021255.11 | Esrrb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Esrrb | mm10_v2_chr12_+_86421628_86421662 | -0.17 | 1.3e-01 | Click! |
| Esrra | mm10_v2_chr19_-_6921753_6921803 | -0.12 | 3.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0021933 | radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 3.7 | 11.2 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 3.3 | 9.9 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) negative regulation of protein import into nucleus, translocation(GO:0033159) regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 3.2 | 9.5 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 2.9 | 8.6 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 2.4 | 7.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 2.3 | 7.0 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 2.3 | 6.8 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 2.2 | 6.7 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 2.1 | 6.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 2.0 | 6.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 2.0 | 6.0 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 1.9 | 17.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.9 | 7.6 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.8 | 8.9 | GO:0019659 | fermentation(GO:0006113) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.7 | 1.7 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 1.6 | 12.6 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.6 | 22.0 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 1.5 | 6.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 1.5 | 4.6 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 1.5 | 13.8 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 1.5 | 19.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 1.5 | 8.8 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 1.4 | 4.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 1.3 | 2.6 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 1.3 | 3.8 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 1.2 | 13.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 1.2 | 4.6 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 1.1 | 5.5 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.1 | 8.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.1 | 4.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.0 | 27.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 1.0 | 9.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 1.0 | 4.9 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.9 | 4.7 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.9 | 2.8 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) |
| 0.9 | 5.6 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.9 | 7.4 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.9 | 2.8 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.9 | 2.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.9 | 5.3 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.8 | 5.9 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.8 | 4.0 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.8 | 2.3 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.8 | 3.8 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.8 | 1.5 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.8 | 5.3 | GO:0090234 | cellular response to testosterone stimulus(GO:0071394) regulation of kinetochore assembly(GO:0090234) |
| 0.7 | 0.7 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.7 | 16.9 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.7 | 2.9 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.7 | 2.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 2.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.7 | 3.5 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.7 | 2.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.6 | 5.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.6 | 1.9 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.6 | 5.0 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.6 | 9.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.6 | 11.8 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.6 | 9.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.6 | 1.9 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.6 | 2.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.6 | 4.9 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.6 | 8.8 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.6 | 3.5 | GO:1902287 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.6 | 2.9 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.6 | 2.8 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.6 | 1.7 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.6 | 3.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.5 | 1.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.5 | 1.6 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 | 19.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.5 | 1.0 | GO:2000823 | regulation of androgen receptor activity(GO:2000823) |
| 0.5 | 2.5 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.5 | 26.7 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.5 | 2.8 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 6.7 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.4 | 3.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.4 | 1.8 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.4 | 1.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 2.9 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.4 | 2.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.4 | 1.7 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.4 | 1.6 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.4 | 3.6 | GO:1900244 | positive regulation of synaptic vesicle endocytosis(GO:1900244) |
| 0.4 | 3.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.4 | 1.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.4 | 1.2 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.4 | 4.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 2.8 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 | 3.5 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.4 | 9.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 1.5 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.4 | 9.9 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.4 | 1.1 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.4 | 2.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 1.1 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.4 | 1.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 2.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.4 | 1.1 | GO:0035672 | regulation of cellular pH reduction(GO:0032847) oligopeptide transmembrane transport(GO:0035672) |
| 0.3 | 5.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 2.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.3 | 3.8 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.3 | 1.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.3 | 11.8 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.3 | 1.0 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.3 | 1.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.3 | 3.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.3 | 0.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.3 | 2.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.3 | 4.3 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.3 | 23.2 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.3 | 18.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.3 | 0.9 | GO:2000612 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) kidney rudiment formation(GO:0072003) kidney field specification(GO:0072004) positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0072108) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) mesenchymal cell apoptotic process involved in metanephros development(GO:1900200) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900211) negative regulation of mesenchymal cell apoptotic process involved in metanephros development(GO:1900212) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) regulation of thyroid hormone generation(GO:2000609) positive regulation of thyroid hormone generation(GO:2000611) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.3 | 5.5 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.3 | 2.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.3 | 6.1 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.3 | 1.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.3 | 2.0 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 0.8 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.3 | 5.0 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.3 | 4.5 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 0.5 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.6 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.3 | 5.8 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.3 | 6.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 1.8 | GO:0061718 | NADH regeneration(GO:0006735) positive regulation of immunoglobulin secretion(GO:0051024) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 2.3 | GO:0035584 | calcium-mediated signaling using intracellular calcium source(GO:0035584) |
| 0.2 | 2.0 | GO:0090336 | response to muscle activity(GO:0014850) positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.2 | 0.7 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.2 | 0.7 | GO:0072384 | organelle transport along microtubule(GO:0072384) |
| 0.2 | 1.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.2 | 1.6 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.2 | 1.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.4 | GO:0048296 | isotype switching to IgA isotypes(GO:0048290) regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 2.0 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.2 | 0.6 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.2 | 0.8 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.2 | 0.6 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.2 | 3.9 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 2.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.2 | 16.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 2.0 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.2 | 3.9 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.2 | 4.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 3.0 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.2 | 3.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.2 | 15.3 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.2 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.1 | GO:0061615 | glycolytic process through fructose-6-phosphate(GO:0061615) |
| 0.2 | 5.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.2 | 0.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.5 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.2 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.2 | 3.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.2 | 0.8 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.2 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.2 | 0.3 | GO:0071649 | negative regulation of mitochondrial fusion(GO:0010637) regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 3.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.5 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.7 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) choline metabolic process(GO:0019695) |
| 0.1 | 5.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 0.6 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.7 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.1 | 0.4 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 3.6 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 1.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.4 | GO:0043691 | high-density lipoprotein particle remodeling(GO:0034375) reverse cholesterol transport(GO:0043691) |
| 0.1 | 0.4 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 4.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 2.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.8 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 1.1 | GO:0032225 | regulation of synaptic transmission, dopaminergic(GO:0032225) |
| 0.1 | 4.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 1.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 3.7 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.3 | GO:0045014 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) detection of glucose(GO:0051594) |
| 0.1 | 1.4 | GO:0042775 | mitochondrial ATP synthesis coupled electron transport(GO:0042775) |
| 0.1 | 0.3 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.1 | 2.4 | GO:2000398 | regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |
| 0.1 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 0.9 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 3.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.6 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.2 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 3.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.9 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 1.5 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.1 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.5 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 4.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.4 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 7.3 | GO:0007612 | learning(GO:0007612) |
| 0.1 | 0.8 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 6.0 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.1 | 0.8 | GO:1990173 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.2 | GO:0046133 | pyrimidine ribonucleoside catabolic process(GO:0046133) |
| 0.1 | 3.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.5 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:2000189 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 2.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 2.3 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.1 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 1.0 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 2.7 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 2.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0002385 | organ or tissue specific immune response(GO:0002251) mucosal immune response(GO:0002385) |
| 0.1 | 0.9 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 3.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 5.1 | GO:0006626 | protein targeting to mitochondrion(GO:0006626) |
| 0.0 | 1.9 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.2 | GO:0030262 | apoptotic nuclear changes(GO:0030262) |
| 0.0 | 1.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.5 | GO:0009311 | oligosaccharide metabolic process(GO:0009311) |
| 0.0 | 0.6 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.8 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.5 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 2.1 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 1.1 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:1903301 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 4.4 | GO:0050808 | synapse organization(GO:0050808) |
| 0.0 | 0.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.9 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 1.8 | GO:0043507 | positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 4.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:1903541 | regulation of exosomal secretion(GO:1903541) positive regulation of exosomal secretion(GO:1903543) |
| 0.0 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 2.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.3 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.7 | GO:0051099 | positive regulation of binding(GO:0051099) |
| 0.0 | 0.9 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.6 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.4 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0098855 | HCN channel complex(GO:0098855) |
| 2.4 | 7.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 1.9 | 28.9 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.7 | 6.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.6 | 12.9 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 1.4 | 14.5 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 1.4 | 22.0 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 1.3 | 4.0 | GO:0005940 | septin ring(GO:0005940) |
| 1.3 | 11.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 1.3 | 2.5 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 1.2 | 7.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 1.2 | 3.6 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 1.1 | 20.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 1.1 | 12.7 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 1.0 | 4.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.0 | 9.5 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.9 | 14.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.8 | 3.8 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.7 | 5.6 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.7 | 9.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.7 | 3.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.6 | 6.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.6 | 3.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.6 | 3.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.6 | 9.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.6 | 1.7 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.5 | 5.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.5 | 3.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 8.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 6.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 1.8 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 1.8 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 1.8 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.4 | 2.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 1.3 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.4 | 19.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.4 | 1.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.4 | 4.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.4 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.4 | 1.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 4.7 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.3 | 5.0 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.3 | 0.6 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.3 | 4.5 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.3 | 7.8 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.3 | 2.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.3 | 3.6 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.3 | 1.1 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.3 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.3 | 2.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.3 | 19.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 35.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.3 | 1.8 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 2.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 5.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 4.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.6 | GO:0098800 | inner mitochondrial membrane protein complex(GO:0098800) |
| 0.2 | 2.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.2 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.2 | 2.5 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.2 | 10.7 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 2.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.2 | 6.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.2 | 9.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 2.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 13.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 10.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 11.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 2.8 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.0 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 3.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 7.1 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 3.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.7 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.1 | 0.7 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.8 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 7.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 4.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.6 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.5 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 8.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.4 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 0.6 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.4 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.1 | 2.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 3.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 4.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 2.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 0.8 | GO:0005832 | zona pellucida receptor complex(GO:0002199) chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 3.5 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 6.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 10.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 9.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.1 | 4.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.2 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 57.2 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.9 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.0 | GO:0044432 | endoplasmic reticulum part(GO:0044432) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.1 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 29.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 3.7 | 11.2 | GO:0031403 | lithium ion binding(GO:0031403) |
| 3.7 | 22.0 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 3.2 | 12.9 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 3.2 | 9.5 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 3.0 | 12.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 2.8 | 19.8 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 2.6 | 12.9 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) |
| 2.3 | 13.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) mercury ion binding(GO:0045340) |
| 2.1 | 10.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 2.0 | 7.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 1.9 | 5.8 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 1.9 | 7.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.8 | 12.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 1.7 | 5.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 1.7 | 5.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 1.5 | 4.6 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 1.5 | 12.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.2 | 8.1 | GO:0015288 | porin activity(GO:0015288) |
| 1.1 | 16.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 1.1 | 18.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.1 | 5.3 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.0 | 6.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 1.0 | 6.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.9 | 20.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.9 | 2.8 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.9 | 2.7 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.9 | 2.7 | GO:0015140 | malate transmembrane transporter activity(GO:0015140) |
| 0.9 | 5.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.9 | 15.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.9 | 3.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.9 | 6.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.9 | 3.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.8 | 7.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.8 | 3.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.8 | 9.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.8 | 3.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.7 | 4.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.7 | 4.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.7 | 4.7 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.7 | 8.6 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.7 | 2.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.7 | 2.0 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.6 | 3.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.6 | 2.5 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.6 | 14.1 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.6 | 1.8 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.6 | 2.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.6 | 1.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.5 | 11.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.5 | 2.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.5 | 3.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.5 | 7.0 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 2.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.5 | 3.4 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.5 | 2.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 4.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.4 | 10.0 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.4 | 8.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 12.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.4 | 3.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 6.4 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.4 | 4.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.4 | 1.7 | GO:0035586 | purinergic receptor activity(GO:0035586) |
| 0.4 | 2.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.4 | 4.1 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.4 | 3.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.4 | 2.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 1.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 6.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 1.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 6.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.3 | 0.9 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.3 | 1.6 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.3 | 0.8 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.3 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.3 | 3.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 2.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 1.6 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.3 | 6.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.3 | 1.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.3 | 2.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 5.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 4.0 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.3 | 3.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 2.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 2.6 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 6.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 4.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 7.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 9.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 6.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 3.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.2 | 0.8 | GO:0004104 | choline kinase activity(GO:0004103) cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 5.0 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 4.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 0.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 0.8 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.2 | 0.6 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.2 | 0.6 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 3.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.2 | 1.9 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 0.6 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.2 | 0.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.2 | 0.7 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.2 | 11.5 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.2 | 0.7 | GO:0051032 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.2 | 38.0 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 2.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 1.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 6.2 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 4.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 7.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 5.0 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.5 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 2.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 1.1 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 0.5 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 0.4 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 1.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 7.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 3.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 3.6 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.6 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 2.6 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 1.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 4.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 2.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.7 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 4.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 2.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.4 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.1 | 3.0 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.1 | 7.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:0036468 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.1 | 3.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 15.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 0.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 4.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.8 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.1 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.7 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.1 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.8 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.6 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 1.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 11.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 0.3 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.1 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.5 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 4.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.5 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.5 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 5.7 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 1.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.1 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 5.2 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.4 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.2 | GO:0004004 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.8 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 22.0 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.8 | 32.9 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.4 | 5.3 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.3 | 3.2 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.2 | 9.5 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.0 | PID_PI3K_PLC_TRK_PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 12.3 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.2 | 22.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 7.7 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.2 | 10.4 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.3 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 11.0 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 2.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.1 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.7 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.8 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 0.7 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.1 | 0.9 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.8 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.8 | PID_P38_ALPHA_BETA_PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 2.0 | PID_HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.7 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.5 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 1.1 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.8 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.0 | 2.1 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID_ERBB1_INTERNALIZATION_PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.2 | PID_BCR_5PATHWAY | BCR signaling pathway |
| 0.0 | 0.5 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.0 | 1.5 | PID_MTOR_4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID_ALPHA_SYNUCLEIN_PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID_ARF6_PATHWAY | Arf6 signaling events |
| 0.0 | 0.3 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID_CXCR4_PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.8 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID_ENDOTHELIN_PATHWAY | Endothelins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 19.8 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 1.8 | 22.0 | REACTOME_ADENYLATE_CYCLASE_ACTIVATING_PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 1.8 | 28.5 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 1.4 | 11.0 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 1.2 | 41.5 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 1.2 | 21.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 1.2 | 15.4 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.6 | 17.5 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.6 | 43.5 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.6 | 12.3 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 7.8 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.5 | 4.3 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 24.4 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.4 | 3.4 | REACTOME_REGULATION_OF_INSULIN_SECRETION_BY_ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 4.1 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 15.6 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.4 | 5.9 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.4 | 15.9 | REACTOME_VOLTAGE_GATED_POTASSIUM_CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.4 | 4.7 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.4 | 9.4 | REACTOME_GLUTATHIONE_CONJUGATION | Genes involved in Glutathione conjugation |
| 0.3 | 7.3 | REACTOME_SIGNAL_ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 3.4 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.3 | 6.8 | REACTOME_PYRUVATE_METABOLISM | Genes involved in Pyruvate metabolism |
| 0.2 | 9.3 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.4 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 16.3 | REACTOME_SIGNALING_BY_NOTCH1 | Genes involved in Signaling by NOTCH1 |
| 0.2 | 3.4 | REACTOME_SYNTHESIS_OF_SUBSTRATES_IN_N_GLYCAN_BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.2 | 12.0 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 2.1 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 6.8 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 2.1 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.6 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 5.8 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.2 | 3.2 | REACTOME_TGF_BETA_RECEPTOR_SIGNALING_IN_EMT_EPITHELIAL_TO_MESENCHYMAL_TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 11.8 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.9 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 5.0 | REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.3 | REACTOME_PYRIMIDINE_CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.5 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 3.2 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 2.6 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 1.8 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.0 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.2 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 2.3 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.1 | 5.5 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.0 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.3 | REACTOME_TETRAHYDROBIOPTERIN_BH4_SYNTHESIS_RECYCLING_SALVAGE_AND_REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 2.0 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.3 | REACTOME_DEADENYLATION_OF_MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 4.7 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.6 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.7 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.1 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.5 | REACTOME_MEMBRANE_BINDING_AND_TARGETTING_OF_GAG_PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.0 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.6 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.3 | REACTOME_CREATION_OF_C4_AND_C2_ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.3 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 2.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME_ENERGY_DEPENDENT_REGULATION_OF_MTOR_BY_LKB1_AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 0.8 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 2.5 | REACTOME_MHC_CLASS_II_ANTIGEN_PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 4.3 | REACTOME_TRANSLATION | Genes involved in Translation |
| 0.0 | 0.9 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.4 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_3_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.8 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.3 | REACTOME_METABOLISM_OF_AMINO_ACIDS_AND_DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME_TRAF6_MEDIATED_INDUCTION_OF_NFKB_AND_MAP_KINASES_UPON_TLR7_8_OR_9_ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 0.3 | REACTOME_CS_DS_DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 2.4 | REACTOME_HOST_INTERACTIONS_OF_HIV_FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.5 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |