Motif ID: Etv3_Erf_Fev_Elk4_Elk1_Elk3
Z-value: 0.661
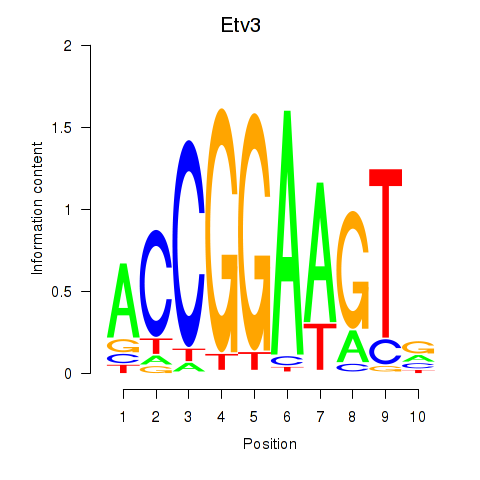
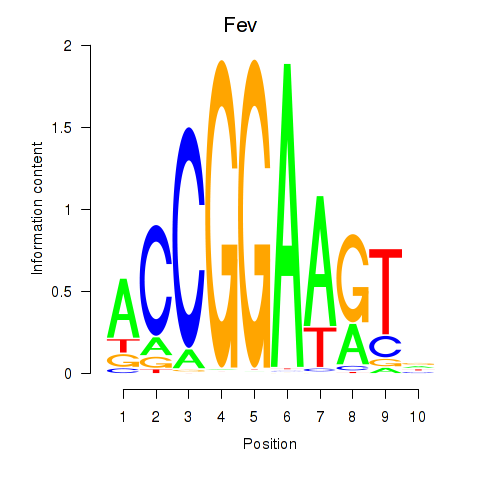
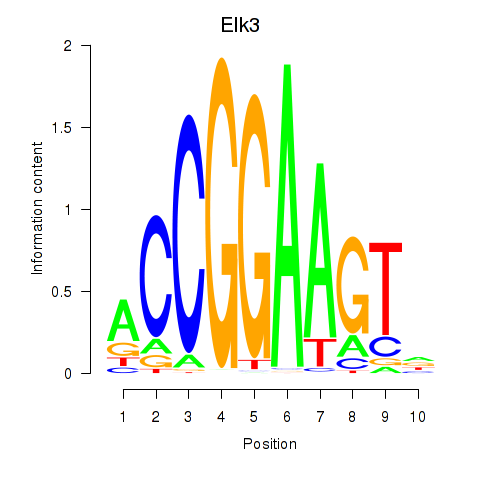
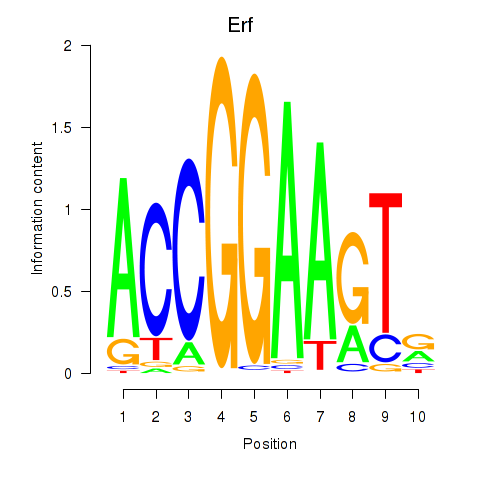
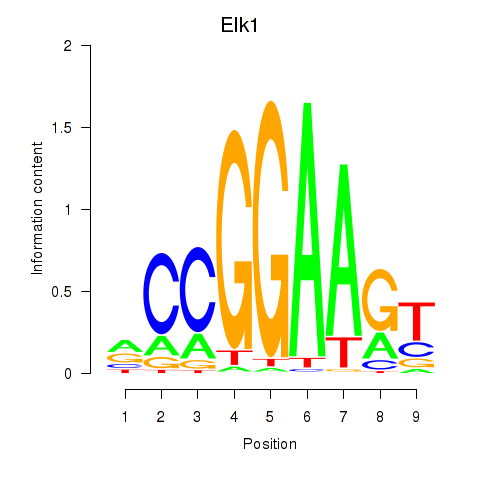
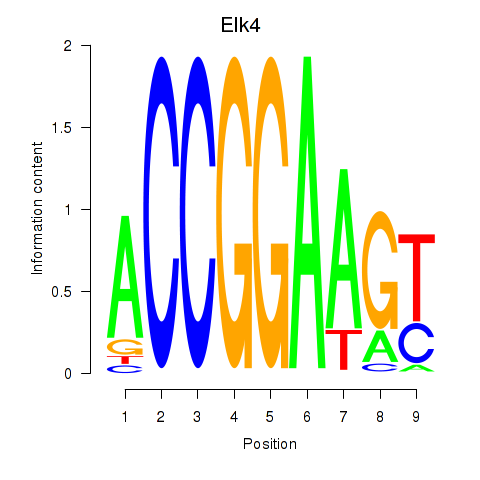
Transcription factors associated with Etv3_Erf_Fev_Elk4_Elk1_Elk3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Elk1 | ENSMUSG00000009406.7 | Elk1 |
| Elk3 | ENSMUSG00000008398.8 | Elk3 |
| Elk4 | ENSMUSG00000026436.9 | Elk4 |
| Erf | ENSMUSG00000040857.9 | Erf |
| Etv3 | ENSMUSG00000003382.12 | Etv3 |
| Fev | ENSMUSG00000055197.4 | Fev |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Elk4 | mm10_v2_chr1_+_132008285_132008316 | 0.58 | 4.2e-08 | Click! |
| Elk3 | mm10_v2_chr10_-_93310963_93310989 | -0.25 | 2.8e-02 | Click! |
| Erf | mm10_v2_chr7_-_25250720_25250761 | 0.18 | 1.1e-01 | Click! |
| Etv3 | mm10_v2_chr3_+_87525572_87525643 | 0.15 | 1.9e-01 | Click! |
| Elk1 | mm10_v2_chrX_-_20950597_20950613 | 0.15 | 2.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 1.5 | 4.4 | GO:1902528 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 1.3 | 3.9 | GO:0019085 | early viral transcription(GO:0019085) |
| 1.1 | 3.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 1.0 | 3.0 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 1.0 | 3.9 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.9 | 4.5 | GO:0021764 | amygdala development(GO:0021764) |
| 0.9 | 3.6 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.9 | 2.6 | GO:0099547 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.8 | 7.6 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.8 | 2.4 | GO:0060003 | elastin biosynthetic process(GO:0051542) copper ion export(GO:0060003) |
| 0.7 | 2.2 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) regulation of microglial cell activation(GO:1903978) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.7 | 2.8 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.7 | 1.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.7 | 2.0 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.6 | 1.9 | GO:2000836 | positive regulation of androgen secretion(GO:2000836) positive regulation of testosterone secretion(GO:2000845) |
| 0.6 | 4.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.6 | 5.6 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.6 | 3.0 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.6 | 2.3 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.5 | 1.6 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.5 | 1.6 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.5 | 1.6 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.5 | 2.6 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.5 | 2.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.5 | 4.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 1.5 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.5 | 1.5 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.5 | 4.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.5 | 0.9 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.5 | 0.5 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.5 | 1.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.4 | 2.1 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.4 | 1.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.4 | 1.6 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.4 | 1.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.4 | 1.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.4 | 1.2 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.4 | 1.1 | GO:0046881 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.4 | 1.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 1.8 | GO:0035617 | stress granule disassembly(GO:0035617) plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.4 | 2.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 1.1 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.4 | 5.7 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 1.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.3 | 1.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.3 | 1.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 7.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.3 | 1.0 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 1.0 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.3 | 1.9 | GO:1903056 | positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 1.0 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.3 | 1.0 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 1.3 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 1.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.3 | 3.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.3 | 1.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 1.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.3 | 1.8 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.3 | 2.1 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.3 | 0.6 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.3 | 0.9 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.3 | 1.7 | GO:0036258 | multivesicular body assembly(GO:0036258) |
| 0.3 | 3.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.3 | 1.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.3 | 0.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.3 | 2.5 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.3 | 1.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.3 | 1.9 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 | 1.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.2 | 0.2 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 3.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.2 | 0.7 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.2 | 0.9 | GO:0021506 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.2 | 1.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 1.9 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.2 | 1.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.2 | 0.7 | GO:0036508 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.2 | 1.6 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.9 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 3.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.6 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 2.0 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.2 | 1.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.2 | 1.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.2 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.0 | GO:0015871 | astrocyte activation involved in immune response(GO:0002265) choline transport(GO:0015871) |
| 0.2 | 3.0 | GO:0003283 | atrial septum development(GO:0003283) |
| 0.2 | 2.0 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.2 | 0.6 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 | 0.9 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.2 | 0.7 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 1.8 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 | 0.5 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.2 | 1.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.2 | 1.7 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.2 | 4.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 3.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.0 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.2 | 0.7 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.2 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 3.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 0.8 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.2 | 7.7 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.2 | 1.7 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.5 | GO:0060295 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.8 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.2 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 1.5 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.2 | 0.5 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.1 | 1.5 | GO:1903351 | response to dopamine(GO:1903350) cellular response to dopamine(GO:1903351) |
| 0.1 | 0.7 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 0.6 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 2.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 3.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.6 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 1.3 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.1 | 0.7 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 2.9 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.1 | 1.0 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.1 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.1 | 0.4 | GO:0097278 | transforming growth factor beta activation(GO:0036363) complement-dependent cytotoxicity(GO:0097278) |
| 0.1 | 0.7 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 1.0 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.4 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.1 | 0.7 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.1 | 0.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 1.3 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.1 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.4 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 0.1 | 0.4 | GO:0061744 | motor behavior(GO:0061744) |
| 0.1 | 1.2 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.4 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.1 | 0.6 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.1 | 2.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.5 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.4 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.1 | 1.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 6.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.8 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.3 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.6 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 | 0.8 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.1 | 0.4 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 2.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 2.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 2.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.1 | 0.9 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.9 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.4 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.6 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.1 | 0.3 | GO:0051892 | regulation of cardioblast differentiation(GO:0051890) negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 1.2 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 1.3 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.8 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.1 | 0.3 | GO:0090327 | negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.4 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 0.8 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.1 | 0.2 | GO:0070488 | regulation of integrin biosynthetic process(GO:0045113) neutrophil aggregation(GO:0070488) |
| 0.1 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:0061357 | Wnt protein secretion(GO:0061355) regulation of Wnt protein secretion(GO:0061356) positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.4 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.1 | 1.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.6 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 1.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.9 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.1 | 1.4 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.2 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.1 | 0.4 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.1 | 1.8 | GO:0051654 | establishment of mitochondrion localization(GO:0051654) |
| 0.1 | 0.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 0.2 | GO:0071649 | regulation of chemokine (C-C motif) ligand 5 production(GO:0071649) |
| 0.1 | 0.2 | GO:1903538 | meiotic sister chromatid cohesion, centromeric(GO:0051754) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.9 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.1 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 1.0 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 0.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.1 | 0.8 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.6 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.1 | 3.3 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.4 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.5 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.1 | 0.4 | GO:1904953 | Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 1.4 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.6 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.4 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.1 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.3 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 1.3 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.2 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 1.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 1.1 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.1 | 0.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 0.2 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.1 | 1.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 3.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.1 | 1.6 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.3 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.6 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.1 | 0.3 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 | 1.3 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 1.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0006272 | DNA replication, synthesis of RNA primer(GO:0006269) leading strand elongation(GO:0006272) |
| 0.1 | 0.3 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 1.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.7 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.0 | 0.7 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.6 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.0 | 1.6 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 2.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.4 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.0 | 1.5 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 0.2 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 1.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 4.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 0.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.8 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.5 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.7 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.4 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.2 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.2 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process(GO:0034627) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.1 | GO:0034398 | telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.2 | GO:0032308 | positive regulation of prostaglandin secretion(GO:0032308) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.6 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 2.5 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0044110 | growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.8 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.3 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 0.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 1.2 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0071883 | regulation of sarcomere organization(GO:0060297) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.7 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.3 | GO:0007188 | adenylate cyclase-modulating G-protein coupled receptor signaling pathway(GO:0007188) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:1903223 | positive regulation of oxidative stress-induced neuron death(GO:1903223) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.4 | GO:0030307 | positive regulation of cell growth(GO:0030307) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.5 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 0.3 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.4 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.0 | 0.2 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.2 | GO:0019221 | cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 | 0.1 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 3.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0050905 | neuromuscular process(GO:0050905) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.2 | GO:0044253 | positive regulation of collagen metabolic process(GO:0010714) positive regulation of multicellular organismal metabolic process(GO:0044253) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0070200 | establishment of protein localization to telomere(GO:0070200) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.0 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 1.8 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 2.7 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0060235 | embryonic retina morphogenesis in camera-type eye(GO:0060059) lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.3 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.2 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 1.2 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 1.8 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 1.2 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.0 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.0 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.0 | GO:1904717 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.0 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.6 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.0 | GO:0070344 | fat cell proliferation(GO:0070341) regulation of fat cell proliferation(GO:0070344) negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 7.6 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 1.1 | 5.6 | GO:1990745 | EARP complex(GO:1990745) |
| 1.1 | 3.3 | GO:0071920 | cleavage body(GO:0071920) |
| 0.8 | 2.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.7 | 5.7 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.7 | 2.8 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.5 | 3.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.5 | 2.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.5 | 3.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.5 | 4.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.5 | 1.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.5 | 1.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.4 | 1.3 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.4 | 3.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 3.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 1.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 1.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 3.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.3 | 2.9 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.3 | 6.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 3.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.3 | 0.9 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.3 | 4.6 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 1.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.3 | 2.2 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.3 | 1.3 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 2.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.2 | 3.7 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.2 | 2.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.2 | 0.7 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.2 | 0.9 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.2 | 1.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.8 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.2 | 0.7 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 2.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 0.6 | GO:0055087 | Ski complex(GO:0055087) |
| 0.2 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 1.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 1.7 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 1.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 1.3 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.2 | 0.9 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 3.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.2 | 1.8 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 3.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 1.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 8.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.2 | 0.6 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 1.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.5 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 3.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.6 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.1 | 0.4 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 1.0 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 2.9 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 1.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.7 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 1.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.1 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.1 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 5.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 1.0 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.8 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.8 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 1.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 0.5 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 8.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 1.3 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.1 | 5.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 6.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.4 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.9 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.1 | 0.2 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 0.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.3 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 0.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.7 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.6 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 2.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 2.3 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 1.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.5 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.0 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 1.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.2 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 3.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 18.1 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 1.1 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 4.6 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 4.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.0 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 9.0 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 0.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 1.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.3 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.9 | 2.6 | GO:0001129 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.8 | 2.4 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.8 | 3.9 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.8 | 4.6 | GO:1904315 | GABA-gated chloride ion channel activity(GO:0022851) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.7 | 2.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.7 | 2.8 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.7 | 4.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.7 | 2.0 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.6 | 2.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.5 | 2.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 2.8 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.4 | 8.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.4 | 7.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.4 | 2.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.4 | 3.3 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.4 | 1.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.4 | 1.2 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.4 | 2.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.4 | 1.6 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 1.6 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.4 | 2.6 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.4 | 1.9 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 2.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 3.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.3 | 2.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 1.0 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.3 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 2.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 0.9 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.3 | 1.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.3 | 1.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.3 | 0.8 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.8 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.3 | 0.8 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.3 | 5.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 1.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 5.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.2 | 1.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.2 | 1.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 1.0 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 5.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 5.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 1.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 2.5 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.2 | 0.9 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 2.0 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.2 | 0.2 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.2 | 0.8 | GO:0035473 | lipase binding(GO:0035473) |
| 0.2 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.6 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.6 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.2 | 2.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.2 | 0.2 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.2 | 2.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.2 | 1.7 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.2 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.0 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 3.8 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.2 | 1.0 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 1.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 1.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 2.1 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.8 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.4 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 2.6 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.9 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.1 | 1.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 3.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.1 | GO:0001179 | RNA polymerase I transcription factor binding(GO:0001179) |
| 0.1 | 4.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 1.6 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 3.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.3 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.1 | 2.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.3 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.4 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 6.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.5 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.4 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 2.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 3.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.6 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 10.5 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.5 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 3.0 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.7 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 4.0 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 2.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 3.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.4 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.1 | 1.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.7 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.0 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.1 | 1.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.9 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 2.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.1 | 0.2 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 0.3 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.4 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 0.4 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.1 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.1 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.6 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.1 | 1.0 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0019961 | interferon binding(GO:0019961) |
| 0.1 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.2 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.5 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.7 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.4 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.2 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.5 | GO:0004407 | histone deacetylase activity(GO:0004407) protein deacetylase activity(GO:0033558) |
| 0.0 | 0.4 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 3.0 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.1 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.0 | 0.4 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 2.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 4.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.7 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.9 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 2.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.9 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.6 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 23.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 4.1 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 2.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.5 | GO:0001191 | transcriptional repressor activity, RNA polymerase II transcription factor binding(GO:0001191) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 2.3 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.4 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 21.7 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 7.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 1.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0031687 | A2A adenosine receptor binding(GO:0031687) |
| 0.0 | 0.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.0 | GO:0016782 | transferase activity, transferring sulfur-containing groups(GO:0016782) sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | SA_G2_AND_M_PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 6.7 | PID_DNA_PK_PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.2 | 4.2 | SA_CASPASE_CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 5.2 | PID_WNT_CANONICAL_PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.3 | SA_G1_AND_S_PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 1.4 | PID_TCR_JNK_PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.0 | PID_NFKAPPAB_CANONICAL_PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 6.0 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 7.9 | SIG_INSULIN_RECEPTOR_PATHWAY_IN_CARDIAC_MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.5 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.1 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.1 | 2.2 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.9 | PID_NCADHERIN_PATHWAY | N-cadherin signaling events |
| 0.1 | 1.6 | ST_GA12_PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.8 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 2.3 | PID_NETRIN_PATHWAY | Netrin-mediated signaling events |
| 0.1 | 2.7 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 2.1 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 1.1 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 1.2 | PID_ARF6_DOWNSTREAM_PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.0 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.3 | PID_TNF_PATHWAY | TNF receptor signaling pathway |
| 0.1 | 1.2 | PID_P38_MKK3_6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.4 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.1 | 0.6 | PID_IL1_PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.0 | 1.1 | PID_EPHB_FWD_PATHWAY | EPHB forward signaling |
| 0.0 | 3.0 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.3 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | ST_INTERFERON_GAMMA_PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.6 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.5 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.0 | 1.9 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.0 | 2.6 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.3 | WNT_SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 3.0 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID_PS1_PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID_TCR_PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID_INTEGRIN_A4B1_PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 1.2 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.1 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | SIG_BCR_SIGNALING_PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.3 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.5 | REACTOME_INTEGRATION_OF_PROVIRUS | Genes involved in Integration of provirus |
| 0.4 | 9.4 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.3 | 5.6 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.3 | 4.6 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 3.4 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.3 | 2.2 | REACTOME_G2_M_DNA_DAMAGE_CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 4.2 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_EARLY_ENDOSOME_MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.3 | 7.1 | REACTOME_NEF_MEDIATES_DOWN_MODULATION_OF_CELL_SURFACE_RECEPTORS_BY_RECRUITING_THEM_TO_CLATHRIN_ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.2 | 2.5 | REACTOME_ACTIVATION_OF_CHAPERONES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.2 | 0.9 | REACTOME_REGULATION_OF_INSULIN_SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.2 | 8.8 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.2 | 3.5 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.2 | 2.4 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 2.2 | REACTOME_COPI_MEDIATED_TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 3.9 | REACTOME_INSULIN_RECEPTOR_RECYCLING | Genes involved in Insulin receptor recycling |
| 0.2 | 3.8 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 0.3 | REACTOME_MRNA_CAPPING | Genes involved in mRNA Capping |
| 0.2 | 0.2 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.2 | 2.9 | REACTOME_CTNNB1_PHOSPHORYLATION_CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.8 | REACTOME_IRAK1_RECRUITS_IKK_COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 5.2 | REACTOME_ION_TRANSPORT_BY_P_TYPE_ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 5.2 | REACTOME_INTRINSIC_PATHWAY_FOR_APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.2 | REACTOME_N_GLYCAN_TRIMMING_IN_THE_ER_AND_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.3 | REACTOME_GENERATION_OF_SECOND_MESSENGER_MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 3.0 | REACTOME_NEP_NS2_INTERACTS_WITH_THE_CELLULAR_EXPORT_MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.1 | 1.6 | REACTOME_GLUTAMATE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 1.2 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.3 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.8 | REACTOME_MITOCHONDRIAL_TRNA_AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.8 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 2.3 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.4 | REACTOME_ADP_SIGNALLING_THROUGH_P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 0.4 | REACTOME_PERK_REGULATED_GENE_EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 1.6 | REACTOME_PROLONGED_ERK_ACTIVATION_EVENTS | Genes involved in Prolonged ERK activation events |
| 0.1 | 1.3 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.1 | 4.4 | REACTOME_REGULATION_OF_WATER_BALANCE_BY_RENAL_AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.7 | REACTOME_RNA_POL_I_TRANSCRIPTION_INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 0.7 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 0.4 | REACTOME_THE_NLRP3_INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.5 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_GOLGI_MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.6 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.4 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.0 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 6.1 | REACTOME_PPARA_ACTIVATES_GENE_EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.1 | 0.5 | REACTOME_PROTEOLYTIC_CLEAVAGE_OF_SNARE_COMPLEX_PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME_EGFR_DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 0.7 | REACTOME_RETROGRADE_NEUROTROPHIN_SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.2 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME_E2F_ENABLED_INHIBITION_OF_PRE_REPLICATION_COMPLEX_FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME_INHIBITION_OF_REPLICATION_INITIATION_OF_DAMAGED_DNA_BY_RB1_E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.8 | REACTOME_GLYCOGEN_BREAKDOWN_GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.0 | REACTOME_CD28_DEPENDENT_PI3K_AKT_SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME_SIGNALING_BY_NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.7 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME_PRE_NOTCH_TRANSCRIPTION_AND_TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.4 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME_DOWNSTREAM_TCR_SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.3 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME_REGULATION_OF_SIGNALING_BY_CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME_G_ALPHA_Z_SIGNALLING_EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME_CHYLOMICRON_MEDIATED_LIPID_TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.7 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME_ERK_MAPK_TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.6 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME_CELL_EXTRACELLULAR_MATRIX_INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME_ACTIVATED_TAK1_MEDIATES_P38_MAPK_ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 1.2 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.3 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.1 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME_NEGATIVE_REGULATORS_OF_RIG_I_MDA5_SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.1 | REACTOME_RNA_POL_I_RNA_POL_III_AND_MITOCHONDRIAL_TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.1 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.3 | REACTOME_GENERIC_TRANSCRIPTION_PATHWAY | Genes involved in Generic Transcription Pathway |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME_METABOLISM_OF_MRNA | Genes involved in Metabolism of mRNA |





