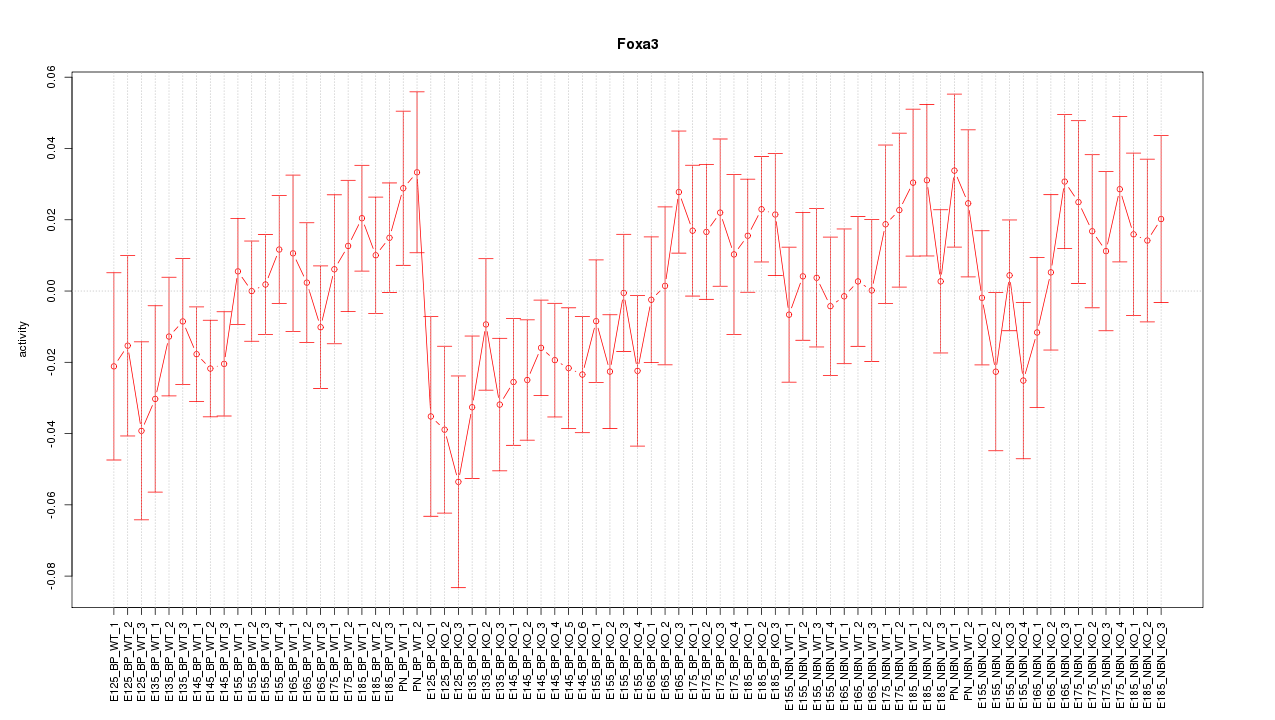
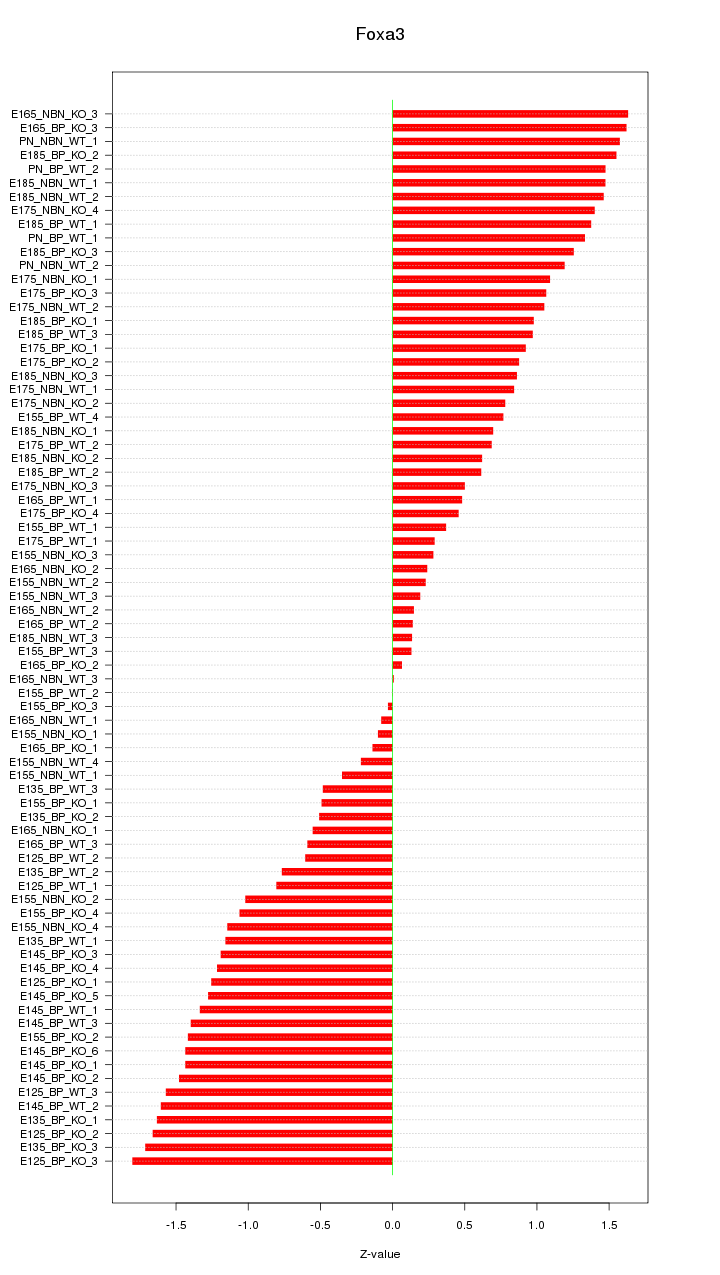
Motif ID: Foxa3
Z-value: 1.024

Transcription factors associated with Foxa3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxa3 | ENSMUSG00000040891.5 | Foxa3 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.7 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 1.8 | 10.8 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 1.7 | 5.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 1.2 | 5.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 1.0 | 7.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 1.0 | 3.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.9 | 2.6 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.8 | 10.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.8 | 7.5 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.8 | 9.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) liver regeneration(GO:0097421) |
| 0.8 | 7.5 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.8 | 14.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 2.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.7 | 2.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.7 | 5.3 | GO:0098707 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.6 | 2.6 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.6 | 3.7 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.6 | 2.4 | GO:0060066 | oviduct development(GO:0060066) |
| 0.6 | 1.7 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.6 | 2.2 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.5 | 2.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.5 | 1.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.5 | 3.9 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.5 | 1.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.5 | 1.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.4 | 1.2 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.4 | 2.3 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.3 | 2.4 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.3 | 4.7 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.3 | 1.9 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.3 | 1.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.3 | 3.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 1.8 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.3 | 2.6 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 | 3.7 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.3 | 7.6 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.3 | 3.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.3 | 1.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 2.9 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 2.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 1.5 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.7 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.2 | 3.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 1.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 3.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 20.0 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.2 | 3.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.8 | GO:1903215 | regulation of mRNA modification(GO:0090365) negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 0.6 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 1.8 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.2 | 4.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.2 | 0.9 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 2.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 7.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.7 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 1.8 | GO:1901028 | regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901028) |
| 0.1 | 1.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 1.5 | GO:2000059 | negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000059) |
| 0.1 | 0.3 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.1 | 10.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 2.6 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.1 | 0.9 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.4 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 2.2 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.4 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.2 | GO:0006553 | lysine metabolic process(GO:0006553) |
| 0.1 | 0.8 | GO:0022027 | interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.1 | 1.1 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 2.1 | GO:0051926 | dopamine receptor signaling pathway(GO:0007212) negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 5.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 6.7 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.7 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.8 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.6 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.7 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.3 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.0 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.3 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 1.6 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.9 | 5.8 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.8 | 9.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.8 | 2.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.8 | 15.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.7 | 5.3 | GO:0097433 | dense body(GO:0097433) |
| 0.4 | 1.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.4 | 2.6 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 2.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 2.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 2.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 2.4 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.2 | 2.3 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 4.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 2.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 5.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 3.9 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 11.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 29.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 5.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 5.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.1 | 0.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 1.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 7.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.6 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 8.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 2.3 | GO:0031968 | mitochondrial outer membrane(GO:0005741) organelle outer membrane(GO:0031968) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 45.3 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 1.8 | 7.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 1.1 | 3.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 1.0 | 5.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.8 | 5.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.6 | 1.9 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.6 | 9.2 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.5 | 2.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.5 | 3.9 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.4 | 1.2 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.4 | 2.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.4 | 1.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 2.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.4 | 2.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 3.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.3 | 1.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.3 | 3.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.3 | 2.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 1.4 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.3 | 10.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 1.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.2 | 3.3 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.2 | 4.1 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 1.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 3.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.2 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.6 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.2 | 0.7 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 2.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 2.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 7.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 2.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 2.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 10.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 4.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.1 | 0.9 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.2 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 6.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 3.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 2.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 2.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 7.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 3.9 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 7.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 1.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 2.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.8 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 3.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.6 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.7 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 5.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 1.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.2 | PID_INTEGRIN5_PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 8.4 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.6 | PID_RHODOPSIN_PATHWAY | Visual signal transduction: Rods |
| 0.2 | 3.7 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.2 | 7.5 | PID_INSULIN_GLUCOSE_PATHWAY | Insulin-mediated glucose transport |
| 0.2 | 2.1 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.2 | 5.8 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.2 | 9.8 | PID_SHP2_PATHWAY | SHP2 signaling |
| 0.1 | 2.9 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.4 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 5.4 | PID_FOXO_PATHWAY | FoxO family signaling |
| 0.1 | 2.1 | PID_IL12_STAT4_PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 2.6 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.1 | 1.3 | PID_ECADHERIN_STABILIZATION_PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 1.6 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.2 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 2.4 | NABA_COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID_IGF1_PATHWAY | IGF1 pathway |
| 0.0 | 1.1 | PID_REG_GR_PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 2.2 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.0 | 3.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.6 | PID_CMYB_PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.8 | PID_P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.5 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | ST_TUMOR_NECROSIS_FACTOR_PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.7 | PID_TRKR_PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID_ECADHERIN_NASCENT_AJ_PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.8 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.7 | 3.7 | REACTOME_OLFACTORY_SIGNALING_PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.4 | 2.8 | REACTOME_APOPTOTIC_CLEAVAGE_OF_CELL_ADHESION_PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 3.6 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 4.6 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.2 | 0.9 | REACTOME_DOWNSTREAM_SIGNAL_TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 2.3 | REACTOME_GABA_A_RECEPTOR_ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 6.0 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.6 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.5 | REACTOME_REGULATED_PROTEOLYSIS_OF_P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.1 | 3.7 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.2 | REACTOME_REGULATION_OF_PYRUVATE_DEHYDROGENASE_PDH_COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 7.1 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.3 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.5 | REACTOME_ACTIVATION_OF_THE_AP1_FAMILY_OF_TRANSCRIPTION_FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 2.1 | REACTOME_PLC_BETA_MEDIATED_EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 2.5 | REACTOME_G1_PHASE | Genes involved in G1 Phase |
| 0.0 | 2.4 | REACTOME_COLLAGEN_FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.5 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.9 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.7 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.9 | REACTOME_G_ALPHA_S_SIGNALLING_EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 1.3 | REACTOME_RESPONSE_TO_ELEVATED_PLATELET_CYTOSOLIC_CA2_ | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |