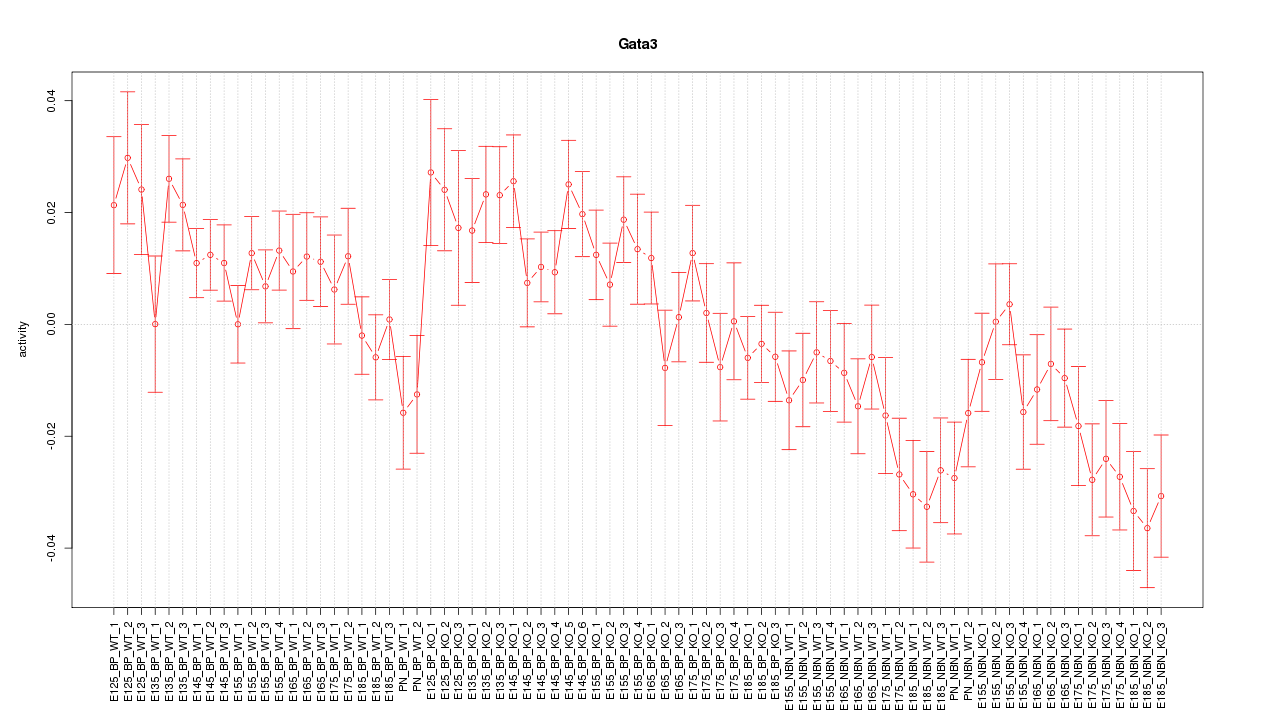
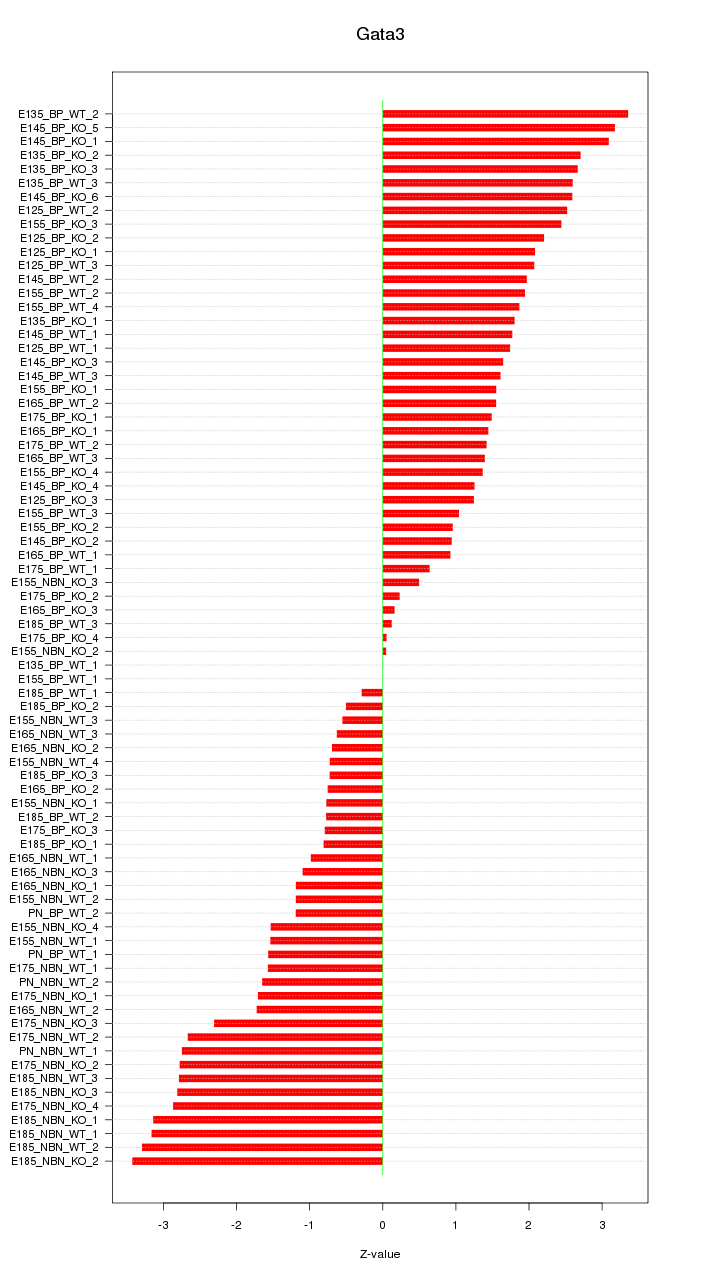
Motif ID: Gata3
Z-value: 1.829

Transcription factors associated with Gata3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gata3 | ENSMUSG00000015619.10 | Gata3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gata3 | mm10_v2_chr2_-_9890026_9890035 | -0.05 | 6.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.8 | 67.0 | GO:0060032 | notochord regression(GO:0060032) |
| 8.9 | 62.4 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 6.5 | 25.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 6.2 | 31.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 6.0 | 18.1 | GO:0001869 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 4.9 | 19.6 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 4.8 | 28.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 4.4 | 48.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 4.4 | 21.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 4.4 | 21.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 4.4 | 13.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 2.9 | 20.5 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 2.8 | 8.4 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 2.8 | 16.7 | GO:0003383 | apical constriction(GO:0003383) |
| 2.7 | 18.7 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 2.5 | 27.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 2.4 | 7.3 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 2.4 | 14.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 2.4 | 28.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 2.3 | 2.3 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 2.3 | 6.9 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.2 | 4.4 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 2.2 | 10.8 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 2.0 | 5.9 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 2.0 | 7.8 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 1.8 | 5.5 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.8 | 5.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 1.8 | 7.0 | GO:0072309 | mesenchymal stem cell maintenance involved in metanephric nephron morphogenesis(GO:0072309) regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 1.7 | 5.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 1.6 | 6.5 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 1.6 | 4.9 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 1.6 | 8.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.6 | 4.7 | GO:0038044 | negative regulation of receptor biosynthetic process(GO:0010871) transforming growth factor-beta secretion(GO:0038044) |
| 1.6 | 6.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 1.5 | 4.6 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 1.5 | 6.0 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 1.4 | 19.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 1.4 | 16.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.3 | 8.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 1.3 | 4.0 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 1.3 | 19.4 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 1.2 | 8.6 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 1.2 | 3.6 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 1.2 | 9.4 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 1.2 | 19.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 1.2 | 4.6 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 1.1 | 18.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.1 | 7.7 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 1.1 | 3.2 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 1.1 | 7.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.1 | 3.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.0 | 5.1 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 1.0 | 1.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.0 | 3.0 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 1.0 | 13.7 | GO:0060013 | righting reflex(GO:0060013) |
| 1.0 | 6.7 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.9 | 5.7 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.9 | 8.4 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.9 | 3.7 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.9 | 6.4 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.9 | 8.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.9 | 7.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.9 | 3.6 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.9 | 3.6 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.9 | 6.3 | GO:1904217 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.9 | 8.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.9 | 2.6 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.8 | 9.3 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.8 | 5.0 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.8 | 3.3 | GO:1903998 | regulation of eating behavior(GO:1903998) |
| 0.8 | 0.8 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.8 | 7.3 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.8 | 5.6 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.8 | 8.0 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.8 | 6.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.8 | 2.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.7 | 2.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.7 | 2.2 | GO:0048162 | preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.7 | 1.4 | GO:0042713 | sperm ejaculation(GO:0042713) |
| 0.7 | 1.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.7 | 5.5 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.7 | 6.7 | GO:0090209 | negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.7 | 3.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.7 | 30.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.6 | 1.9 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.6 | 1.3 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.6 | 3.2 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.6 | 1.3 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.6 | 0.6 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.6 | 16.2 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.6 | 1.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.6 | 4.9 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.6 | 11.1 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.6 | 17.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.6 | 1.8 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.6 | 1.2 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.6 | 2.9 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.6 | 1.7 | GO:0036233 | glycine import(GO:0036233) |
| 0.6 | 1.7 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.6 | 10.0 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.5 | 1.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.5 | 3.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.5 | 1.1 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.5 | 4.3 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.5 | 2.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.5 | 2.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.5 | 2.7 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 2.1 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.5 | 1.0 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.5 | 1.5 | GO:0016056 | photoreceptor cell morphogenesis(GO:0008594) rhodopsin mediated signaling pathway(GO:0016056) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.5 | 2.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.5 | 3.1 | GO:0007276 | gamete generation(GO:0007276) |
| 0.5 | 1.5 | GO:0035864 | response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.5 | 9.1 | GO:0051450 | myoblast proliferation(GO:0051450) |
| 0.5 | 2.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.5 | 3.5 | GO:0072224 | metanephric glomerulus development(GO:0072224) |
| 0.5 | 3.5 | GO:0089700 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) protein kinase D signaling(GO:0089700) |
| 0.5 | 8.2 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.5 | 1.9 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.5 | 14.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 0.9 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.5 | 5.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.5 | 1.4 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.5 | 1.4 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) positive regulation of aminoacyl-tRNA ligase activity(GO:1903632) regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.5 | 1.4 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.5 | 2.7 | GO:0051461 | regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.4 | 1.3 | GO:0097350 | neutrophil clearance(GO:0097350) |
| 0.4 | 3.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.4 | 2.6 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) |
| 0.4 | 5.2 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.4 | 0.4 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.4 | 2.6 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.4 | 3.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.4 | 1.3 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.4 | 3.3 | GO:1902730 | positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.4 | 2.0 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.4 | 1.2 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.4 | 1.2 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.4 | 1.2 | GO:1990927 | negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 3.8 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.4 | 1.2 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.4 | 1.9 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.4 | 1.1 | GO:2000373 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.4 | 0.7 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.4 | 1.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.4 | 2.9 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.4 | 6.6 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.4 | 3.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.4 | 2.1 | GO:0003263 | cardioblast proliferation(GO:0003263) regulation of cardioblast proliferation(GO:0003264) |
| 0.3 | 2.1 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.3 | 1.3 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.3 | 2.0 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.3 | 5.7 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.3 | 1.7 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.3 | 1.0 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.3 | 3.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.3 | 1.9 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.3 | 1.6 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 | 2.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.3 | 2.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 1.2 | GO:0019230 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.3 | 4.7 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 2.5 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.3 | 1.2 | GO:0090298 | mitochondrial DNA repair(GO:0043504) negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.3 | 1.8 | GO:2001185 | regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.3 | 2.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 4.2 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.3 | 1.7 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.3 | 5.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.3 | 6.9 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 5.4 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.3 | 1.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.3 | 5.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 1.4 | GO:0050912 | detection of chemical stimulus involved in sensory perception(GO:0050907) detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.3 | 4.7 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.3 | 9.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.3 | 1.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.3 | 3.9 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.3 | 2.9 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.3 | 8.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) |
| 0.3 | 3.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.3 | 1.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.3 | 0.8 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.3 | 2.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 | 7.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.3 | 0.5 | GO:2000157 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.3 | 1.0 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.2 | 1.0 | GO:0061101 | neuroendocrine cell differentiation(GO:0061101) |
| 0.2 | 1.7 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 2.9 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 | 1.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 1.2 | GO:0038030 | non-canonical Wnt signaling pathway via MAPK cascade(GO:0038030) non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.2 | 4.6 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.2 | 3.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 0.9 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.2 | 16.6 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.2 | 1.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.2 | 1.6 | GO:2000254 | regulation of male germ cell proliferation(GO:2000254) |
| 0.2 | 4.9 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.2 | 0.9 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.2 | 0.2 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.5 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 6.4 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.2 | 1.5 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.2 | 1.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 | 0.8 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 1.6 | GO:0010835 | regulation of protein ADP-ribosylation(GO:0010835) |
| 0.2 | 2.0 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.2 | 1.0 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.2 | 1.5 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 0.4 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 4.5 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 3.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.2 | 2.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 1.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 4.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.2 | 0.7 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 0.7 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 1.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.2 | 1.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.2 | 3.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 1.5 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 1.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 1.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 1.0 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.2 | 1.4 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.2 | 1.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.2 | 0.8 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.2 | 0.6 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 0.8 | GO:0061038 | uterus morphogenesis(GO:0061038) |
| 0.2 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 1.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 2.7 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 3.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.1 | 1.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.7 | GO:0032329 | serine transport(GO:0032329) |
| 0.1 | 4.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.1 | 0.4 | GO:0006667 | sphinganine metabolic process(GO:0006667) regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.1 | 1.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 | 3.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 3.1 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.1 | 0.4 | GO:0071380 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.5 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) |
| 0.1 | 1.2 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 3.8 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.1 | 5.0 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.1 | 0.6 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 1.5 | GO:1903504 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 2.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 1.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 1.4 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.8 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.2 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 2.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.1 | 4.1 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 1.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 1.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.4 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) |
| 0.1 | 1.6 | GO:0046622 | positive regulation of organ growth(GO:0046622) |
| 0.1 | 3.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.7 | GO:0098703 | parathyroid hormone secretion(GO:0035898) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 3.5 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 4.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.1 | 1.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.3 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.6 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 3.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.3 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 8.0 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 1.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 1.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.1 | 3.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.6 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.6 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 3.7 | GO:0010634 | positive regulation of epithelial cell migration(GO:0010634) |
| 0.1 | 1.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 2.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 2.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.9 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 1.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 1.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 4.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.6 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.6 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.1 | 0.1 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.7 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 1.9 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.1 | 6.3 | GO:0006413 | translational initiation(GO:0006413) |
| 0.1 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.5 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.1 | 0.9 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.6 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.1 | 1.0 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.1 | 1.0 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 1.7 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 0.7 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.3 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.6 | GO:0009583 | detection of light stimulus(GO:0009583) |
| 0.1 | 1.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.1 | 1.9 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.1 | 0.8 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.3 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.7 | GO:1903313 | positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.2 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 1.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.1 | GO:0048854 | brain morphogenesis(GO:0048854) |
| 0.0 | 3.3 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.0 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.8 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.7 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 6.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.4 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.1 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.1 | GO:0035330 | regulation of hippo signaling(GO:0035330) |
| 0.0 | 0.1 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.0 | 0.7 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.4 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.9 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 1.0 | GO:1901184 | regulation of ERBB signaling pathway(GO:1901184) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 1.1 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 1.5 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.0 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.4 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 1.1 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.4 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 1.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.7 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.3 | GO:0090132 | epithelial cell migration(GO:0010631) tissue migration(GO:0090130) epithelium migration(GO:0090132) |
| 0.0 | 0.5 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 1.7 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.7 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.2 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.6 | GO:0009953 | dorsal/ventral pattern formation(GO:0009953) |
| 0.0 | 0.1 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.4 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 1.1 | GO:0048515 | spermatid differentiation(GO:0048515) |
| 0.0 | 0.1 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.7 | GO:1903749 | positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.3 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.2 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 67.7 | GO:0097542 | ciliary tip(GO:0097542) |
| 4.9 | 9.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 2.7 | 8.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 2.6 | 13.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 2.0 | 6.0 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 2.0 | 6.0 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 2.0 | 8.0 | GO:0090537 | CERF complex(GO:0090537) |
| 1.8 | 5.5 | GO:0071914 | prominosome(GO:0071914) |
| 1.8 | 14.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.7 | 5.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.7 | 8.5 | GO:0031523 | Myb complex(GO:0031523) |
| 1.6 | 4.7 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 1.2 | 7.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 1.2 | 3.6 | GO:0031088 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 1.1 | 4.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 1.1 | 3.2 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 1.0 | 6.2 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 1.0 | 15.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 1.0 | 3.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 1.0 | 4.9 | GO:0033648 | host intracellular organelle(GO:0033647) host intracellular membrane-bounded organelle(GO:0033648) |
| 0.9 | 3.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 3.7 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.8 | 3.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.8 | 30.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.8 | 18.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.7 | 7.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.7 | 2.7 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.7 | 20.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.6 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 3.5 | GO:0043256 | laminin complex(GO:0043256) |
| 0.6 | 6.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.6 | 7.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.6 | 26.0 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.6 | 5.0 | GO:0002177 | manchette(GO:0002177) |
| 0.6 | 1.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.5 | 5.3 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.5 | 3.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.5 | 2.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 2.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.5 | 4.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.5 | 2.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 2.0 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.5 | 1.9 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.5 | 2.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.5 | 6.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 2.8 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.5 | 10.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.5 | 1.4 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.4 | 2.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.4 | 7.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 0.8 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.4 | 3.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.4 | 16.4 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 1.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 2.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.4 | 1.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.4 | 2.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.4 | 4.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.3 | 1.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.3 | 4.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.3 | 1.3 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 2.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.3 | 3.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 3.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.3 | 2.0 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.3 | 9.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.3 | 16.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 1.9 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.3 | 2.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.3 | 0.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.3 | 1.8 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.5 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 3.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.2 | 32.4 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.2 | 1.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 1.6 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 1.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.2 | 7.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 1.9 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 7.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 1.5 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.2 | 3.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 1.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.2 | 1.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 6.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.2 | 1.7 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.2 | 1.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.2 | 9.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 2.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.2 | 4.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.2 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 12.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.3 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 1.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 50.7 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 1.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.6 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 6.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 4.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 4.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.2 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.9 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 6.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.6 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.1 | 1.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.4 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 4.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 1.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 4.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.7 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 7.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 11.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 2.6 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.0 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 2.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.4 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.1 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 4.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.3 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.1 | 1.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.1 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.5 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.7 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 4.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 4.0 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 6.7 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 3.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.9 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 4.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 13.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 159.1 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 1.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 15.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 4.7 | 4.7 | GO:0001846 | opsonin binding(GO:0001846) |
| 4.4 | 21.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 3.7 | 18.4 | GO:0043532 | angiostatin binding(GO:0043532) |
| 3.6 | 10.8 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 3.2 | 19.4 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 2.4 | 7.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 2.3 | 6.9 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 2.2 | 8.7 | GO:0008142 | oxysterol binding(GO:0008142) |
| 2.0 | 72.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 1.9 | 32.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.9 | 5.6 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 1.8 | 16.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 1.8 | 5.4 | GO:0016015 | morphogen activity(GO:0016015) |
| 1.8 | 5.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.7 | 8.3 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 1.6 | 4.9 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.6 | 7.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 1.5 | 4.6 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 1.5 | 4.6 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 1.5 | 4.5 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 1.5 | 14.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.3 | 6.5 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 1.3 | 22.7 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 1.2 | 6.0 | GO:0035240 | dopamine binding(GO:0035240) |
| 1.1 | 24.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.1 | 1.1 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) |
| 1.1 | 3.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 1.1 | 4.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 1.1 | 16.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 1.1 | 3.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 1.1 | 3.2 | GO:0036004 | GAF domain binding(GO:0036004) |
| 1.0 | 5.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 1.0 | 4.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 2.0 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 1.0 | 18.1 | GO:0001848 | complement binding(GO:0001848) |
| 0.9 | 6.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.9 | 6.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.9 | 4.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.9 | 10.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.9 | 2.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.8 | 5.0 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.8 | 3.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.8 | 4.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.8 | 3.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.8 | 5.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.8 | 0.8 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.8 | 2.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.7 | 2.9 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.7 | 2.9 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.7 | 2.9 | GO:0043176 | amine binding(GO:0043176) |
| 0.7 | 7.0 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.7 | 1.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.7 | 6.8 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 3.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.6 | 1.9 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.6 | 7.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.6 | 3.6 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.6 | 3.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.6 | 4.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.6 | 8.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.5 | 1.6 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.5 | 2.2 | GO:0030292 | hyalurononglucosaminidase activity(GO:0004415) protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 2.7 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 3.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.5 | 2.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 4.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.5 | 97.0 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.5 | 0.5 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.5 | 5.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.5 | 3.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.5 | 1.4 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.5 | 4.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.5 | 4.6 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.5 | 1.4 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.4 | 6.3 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 3.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.4 | 7.4 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.4 | 6.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.4 | 1.3 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.4 | 6.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.4 | 15.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.4 | 10.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.4 | 5.7 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.4 | 1.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 17.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.4 | 6.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.4 | 1.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.4 | 10.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 1.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 5.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.4 | 10.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.4 | 9.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 3.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.4 | 1.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.4 | 15.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 5.0 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 1.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.3 | 1.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.3 | 4.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 10.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.3 | 3.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 2.6 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.3 | 1.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 0.9 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 28.6 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.3 | 3.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 1.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 14.2 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.3 | 3.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 1.7 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.3 | 1.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.3 | 5.8 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.3 | 1.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 4.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 5.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 3.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.3 | 8.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.3 | 2.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.3 | 1.3 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 3.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 1.5 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.3 | 1.0 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 10.7 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.2 | 3.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 5.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.2 | 7.5 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.2 | 2.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.8 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.2 | 1.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 38.3 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.2 | 2.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 4.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 9.0 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.2 | 1.7 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 0.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.2 | 1.0 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.2 | 1.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 8.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.2 | 0.6 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 1.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 1.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 3.8 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.2 | 1.4 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.2 | 1.7 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.2 | 1.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 5.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.2 | 0.6 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.2 | 0.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.2 | 3.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.2 | 41.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.2 | 1.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 1.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 6.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 0.5 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.2 | 1.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.8 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.2 | 5.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.2 | 1.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 1.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.2 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 1.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 6.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 5.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.1 | 0.4 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 21.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.7 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 1.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 2.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 1.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 5.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 2.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.1 | 1.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 4.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 1.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.1 | 0.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.9 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.3 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 6.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.1 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 2.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 10.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 1.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 3.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.4 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 2.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 2.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 10.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.7 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.0 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.4 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 14.1 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 1.6 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.8 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 2.1 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0019103 | pyrimidine nucleotide binding(GO:0019103) |
| 0.0 | 0.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 1.8 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.0 | 0.1 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.8 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.8 | GO:0043178 | alcohol binding(GO:0043178) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 72.7 | PID_HEDGEHOG_2PATHWAY | Signaling events mediated by the Hedgehog family |
| 1.0 | 48.5 | PID_AURORA_B_PATHWAY | Aurora B signaling |
| 0.9 | 27.5 | PID_WNT_SIGNALING_PATHWAY | Wnt signaling network |
| 0.6 | 27.0 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.6 | 12.3 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.5 | 1.9 | ST_IL_13_PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.4 | 3.5 | PID_INTEGRIN4_PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.4 | 3.4 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.4 | 4.0 | PID_ALK2_PATHWAY | ALK2 signaling events |
| 0.3 | 11.5 | PID_P38_MK2_PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.3 | 8.6 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.3 | 6.8 | PID_HDAC_CLASSIII_PATHWAY | Signaling events mediated by HDAC Class III |
| 0.3 | 7.7 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.3 | 24.4 | PID_E2F_PATHWAY | E2F transcription factor network |
| 0.3 | 21.7 | SIG_PIP3_SIGNALING_IN_CARDIAC_MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.3 | 16.0 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 11.7 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.3 | 5.3 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.3 | 17.3 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.2 | 4.6 | PID_HIV_NEF_PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.2 | 3.2 | PID_ERB_GENOMIC_PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 2.6 | PID_CD40_PATHWAY | CD40/CD40L signaling |
| 0.2 | 1.7 | PID_IL5_PATHWAY | IL5-mediated signaling events |
| 0.2 | 1.0 | SA_B_CELL_RECEPTOR_COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.2 | 7.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 3.9 | ST_JNK_MAPK_PATHWAY | JNK MAPK Pathway |
| 0.1 | 0.7 | PID_PTP1B_PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 4.6 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.1 | 0.9 | PID_IL2_STAT5_PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.1 | 17.0 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 4.0 | PID_RAC1_REG_PATHWAY | Regulation of RAC1 activity |
| 0.1 | 7.6 | PID_AR_PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.0 | PID_ECADHERIN_KERATINOCYTE_PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.1 | PID_EPHRINB_REV_PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.0 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.1 | 1.2 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.2 | ST_WNT_BETA_CATENIN_PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.5 | PID_ERA_GENOMIC_PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 3.2 | PID_HES_HEY_PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.1 | PID_P53_DOWNSTREAM_PATHWAY | Direct p53 effectors |
| 0.1 | 1.9 | PID_ATM_PATHWAY | ATM pathway |
| 0.1 | 2.1 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 1.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.7 | PID_PLK1_PATHWAY | PLK1 signaling events |
| 0.0 | 4.6 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID_FANCONI_PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID_PI3KCI_AKT_PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID_ERBB1_DOWNSTREAM_PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.4 | PID_ILK_PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 6.9 | REACTOME_ETHANOL_OXIDATION | Genes involved in Ethanol oxidation |
| 1.7 | 17.1 | REACTOME_INTRINSIC_PATHWAY | Genes involved in Intrinsic Pathway |
| 1.5 | 40.1 | REACTOME_KINESINS | Genes involved in Kinesins |
| 0.8 | 24.1 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.6 | 20.0 | REACTOME_TIGHT_JUNCTION_INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 7.2 | REACTOME_IL_6_SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.6 | 15.2 | REACTOME_G0_AND_EARLY_G1 | Genes involved in G0 and Early G1 |
| 0.6 | 7.8 | REACTOME_NOTCH_HLH_TRANSCRIPTION_PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.6 | 14.5 | REACTOME_REGULATION_OF_GENE_EXPRESSION_IN_BETA_CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.5 | 6.0 | REACTOME_AMINE_DERIVED_HORMONES | Genes involved in Amine-derived hormones |
| 0.5 | 13.8 | REACTOME_YAP1_AND_WWTR1_TAZ_STIMULATED_GENE_EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.4 | 6.0 | REACTOME_P38MAPK_EVENTS | Genes involved in p38MAPK events |
| 0.4 | 4.7 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.4 | 2.3 | REACTOME_TRANSLOCATION_OF_ZAP_70_TO_IMMUNOLOGICAL_SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.4 | 13.0 | REACTOME_FATTY_ACYL_COA_BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.4 | 6.3 | REACTOME_ACTIVATION_OF_BH3_ONLY_PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.4 | 11.0 | REACTOME_IL_RECEPTOR_SHC_SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.3 | 7.3 | REACTOME_CITRIC_ACID_CYCLE_TCA_CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 5.0 | REACTOME_HYALURONAN_METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.3 | 11.8 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.3 | 18.8 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 5.0 | REACTOME_MITOCHONDRIAL_FATTY_ACID_BETA_OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 2.6 | REACTOME_DOWNREGULATION_OF_ERBB2_ERBB3_SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 13.7 | REACTOME_RNA_POL_I_PROMOTER_OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 4.0 | REACTOME_DEGRADATION_OF_THE_EXTRACELLULAR_MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.3 | 2.5 | REACTOME_METABOLISM_OF_MRNA | Genes involved in Metabolism of mRNA |
| 0.2 | 2.1 | REACTOME_INTERACTIONS_OF_VPR_WITH_HOST_CELLULAR_PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 4.6 | REACTOME_ABCA_TRANSPORTERS_IN_LIPID_HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 2.1 | REACTOME_CDC6_ASSOCIATION_WITH_THE_ORC_ORIGIN_COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.2 | 2.2 | REACTOME_INHIBITION_OF_INSULIN_SECRETION_BY_ADRENALINE_NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.2 | 3.7 | REACTOME_VIRAL_MESSENGER_RNA_SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 6.6 | REACTOME_SEMA4D_INDUCED_CELL_MIGRATION_AND_GROWTH_CONE_COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.2 | 3.5 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.2 | 4.1 | REACTOME_DCC_MEDIATED_ATTRACTIVE_SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.2 | 14.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.2 | 0.4 | REACTOME_SHC1_EVENTS_IN_EGFR_SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.2 | 14.1 | REACTOME_G_ALPHA1213_SIGNALLING_EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.2 | 5.4 | REACTOME_PIP3_ACTIVATES_AKT_SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.2 | 15.3 | REACTOME_CLASS_B_2_SECRETIN_FAMILY_RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.2 | 4.1 | REACTOME_MEIOTIC_RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 1.9 | REACTOME_SEMA3A_PLEXIN_REPULSION_SIGNALING_BY_INHIBITING_INTEGRIN_ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 3.4 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 4.2 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.1 | 3.3 | REACTOME_RNA_POL_III_TRANSCRIPTION_INITIATION_FROM_TYPE_2_PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 1.2 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 9.2 | REACTOME_APC_C_CDH1_MEDIATED_DEGRADATION_OF_CDC20_AND_OTHER_APC_C_CDH1_TARGETED_PROTEINS_IN_LATE_MITOSIS_EARLY_G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.5 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.1 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 3.8 | REACTOME_IRON_UPTAKE_AND_TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 2.5 | REACTOME_METABOLISM_OF_PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.7 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.8 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 2.9 | REACTOME_STRIATED_MUSCLE_CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.2 | REACTOME_PKB_MEDIATED_EVENTS | Genes involved in PKB-mediated events |
| 0.1 | 8.0 | REACTOME_ANTIVIRAL_MECHANISM_BY_IFN_STIMULATED_GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 18.4 | REACTOME_ANTIGEN_PROCESSING_UBIQUITINATION_PROTEASOME_DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.7 | REACTOME_TRAF6_MEDIATED_IRF7_ACTIVATION_IN_TLR7_8_OR_9_SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.8 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 8.1 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 0.4 | REACTOME_HOMOLOGOUS_RECOMBINATION_REPAIR_OF_REPLICATION_INDEPENDENT_DOUBLE_STRAND_BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.9 | REACTOME_PRE_NOTCH_EXPRESSION_AND_PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.1 | 1.5 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.9 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 4.8 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.9 | REACTOME_AMINO_ACID_SYNTHESIS_AND_INTERCONVERSION_TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.2 | REACTOME_FANCONI_ANEMIA_PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.9 | REACTOME_RESOLUTION_OF_AP_SITES_VIA_THE_SINGLE_NUCLEOTIDE_REPLACEMENT_PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 1.9 | REACTOME_REGULATION_OF_MRNA_STABILITY_BY_PROTEINS_THAT_BIND_AU_RICH_ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 0.7 | REACTOME_ACTIVATION_OF_NF_KAPPAB_IN_B_CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 0.5 | REACTOME_SIGNALING_BY_FGFR3_MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.8 | REACTOME_JNK_C_JUN_KINASES_PHOSPHORYLATION_AND_ACTIVATION_MEDIATED_BY_ACTIVATED_HUMAN_TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 1.0 | REACTOME_IONOTROPIC_ACTIVITY_OF_KAINATE_RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME_MRNA_DECAY_BY_5_TO_3_EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.5 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME_DOWNREGULATION_OF_SMAD2_3_SMAD4_TRANSCRIPTIONAL_ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 3.2 | REACTOME_FACTORS_INVOLVED_IN_MEGAKARYOCYTE_DEVELOPMENT_AND_PLATELET_PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 3.4 | REACTOME_SIGNALING_BY_RHO_GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME_PURINE_CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.5 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME_GAP_JUNCTION_ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.2 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.4 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.1 | REACTOME_REGULATION_OF_IFNG_SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |