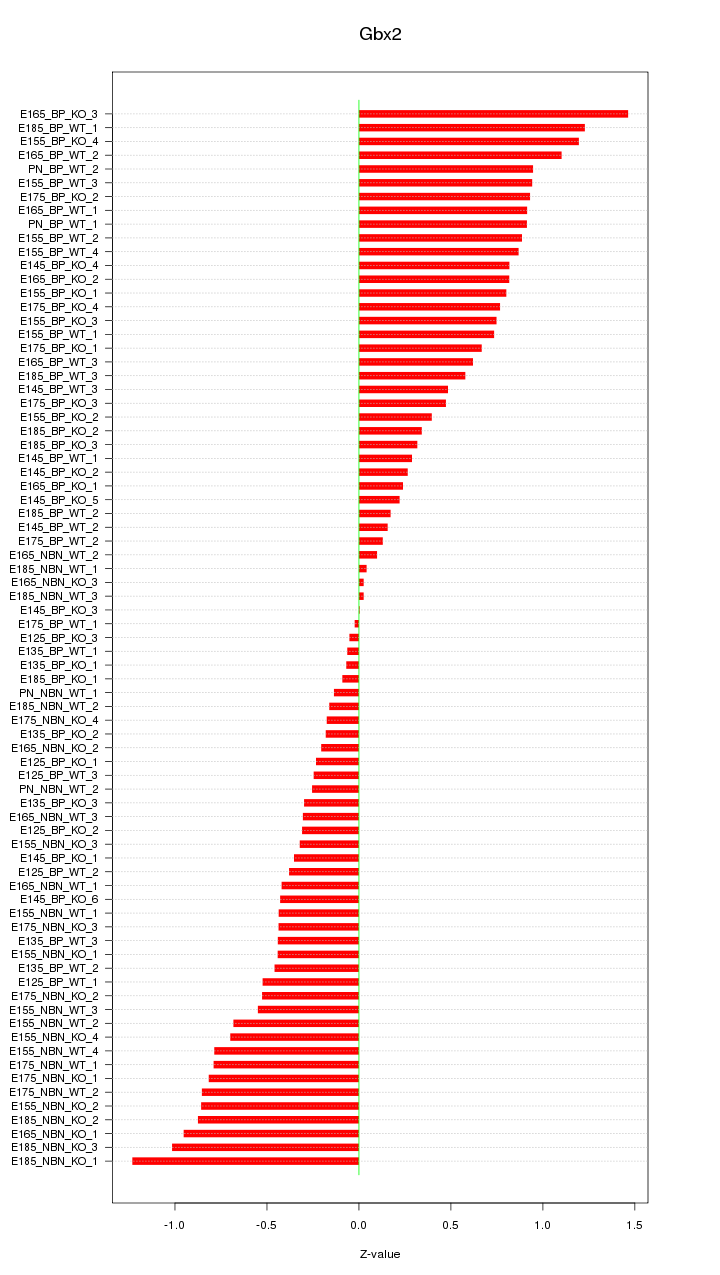
Motif ID: Gbx2
Z-value: 0.623
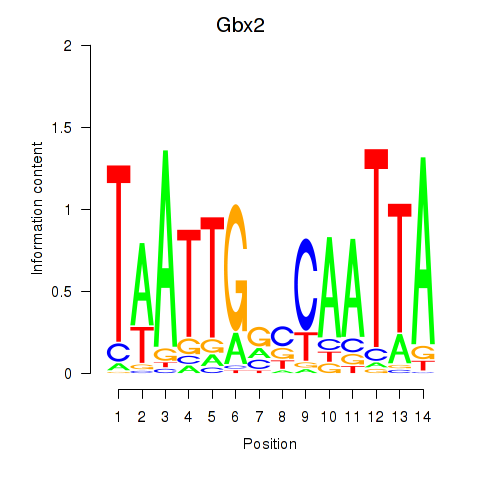
Transcription factors associated with Gbx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gbx2 | ENSMUSG00000034486.7 | Gbx2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gbx2 | mm10_v2_chr1_-_89933290_89933290 | -0.08 | 4.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 17.5 | GO:0042891 | antibiotic transport(GO:0042891) dipeptide transport(GO:0042938) |
| 1.9 | 7.8 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 1.1 | 4.3 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.0 | 2.9 | GO:0099548 | drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.9 | 2.8 | GO:2000564 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) positive regulation of CD8-positive, alpha-beta T cell activation(GO:2001187) |
| 0.6 | 3.8 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.6 | 4.9 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.6 | 1.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.5 | 4.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.4 | 2.6 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.4 | 4.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.3 | 1.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 3.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.1 | 0.4 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.1 | 1.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.7 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 4.6 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.1 | 0.3 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.0 | 0.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.3 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.4 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 2.3 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 1.4 | GO:0042742 | defense response to bacterium(GO:0042742) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 2.9 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 2.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 8.8 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 7.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 3.0 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 2.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 3.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.8 | 17.5 | GO:0042936 | dipeptide transporter activity(GO:0042936) |
| 1.5 | 4.6 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 1.2 | 4.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 1.2 | 4.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 2.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.5 | 3.8 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 3.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.4 | 7.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 1.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 2.8 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.3 | 2.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.7 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 4.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.1 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 2.9 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 0.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.4 | GO:0015087 | cobalt ion transmembrane transporter activity(GO:0015087) |
| 0.0 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 4.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 1.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 2.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | PID_INTEGRIN2_PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 3.1 | PID_LPA4_PATHWAY | LPA4-mediated signaling events |
| 0.1 | 6.7 | PID_RHOA_REG_PATHWAY | Regulation of RhoA activity |
| 0.1 | 1.7 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID_INTEGRIN_CS_PATHWAY | Integrin family cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME_P2Y_RECEPTORS | Genes involved in P2Y receptors |
| 0.3 | 17.5 | REACTOME_AMINO_ACID_AND_OLIGOPEPTIDE_SLC_TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.2 | 4.7 | REACTOME_IMMUNOREGULATORY_INTERACTIONS_BETWEEN_A_LYMPHOID_AND_A_NON_LYMPHOID_CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 2.3 | REACTOME_APOPTOSIS_INDUCED_DNA_FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.9 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 6.7 | REACTOME_NRAGE_SIGNALS_DEATH_THROUGH_JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 4.9 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME_PRESYNAPTIC_NICOTINIC_ACETYLCHOLINE_RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME_REMOVAL_OF_THE_FLAP_INTERMEDIATE_FROM_THE_C_STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.0 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |