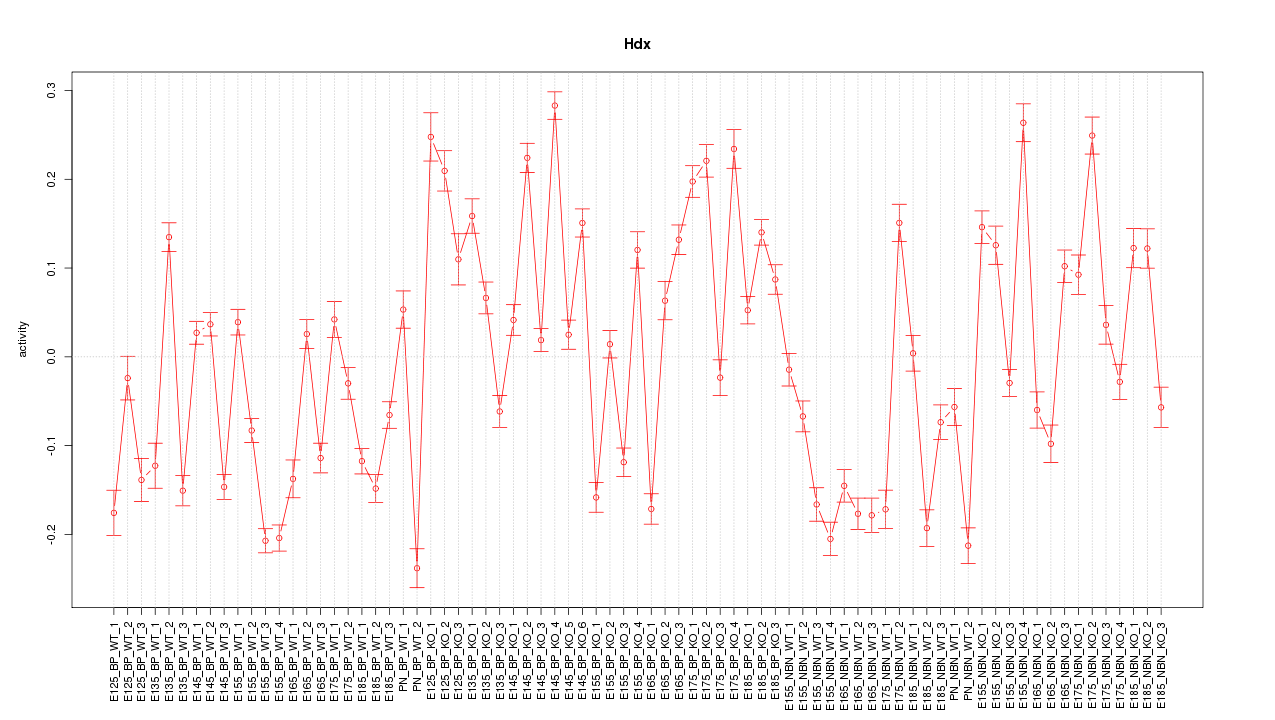
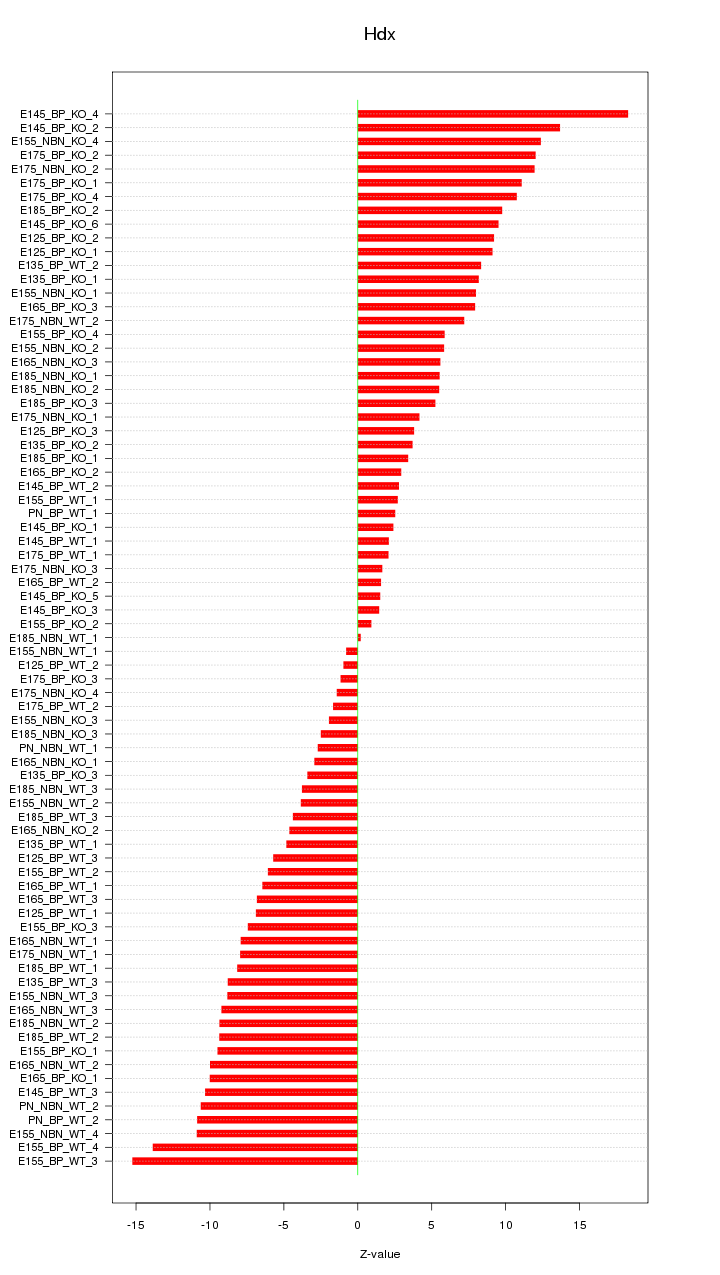
Motif ID: Hdx
Z-value: 7.518
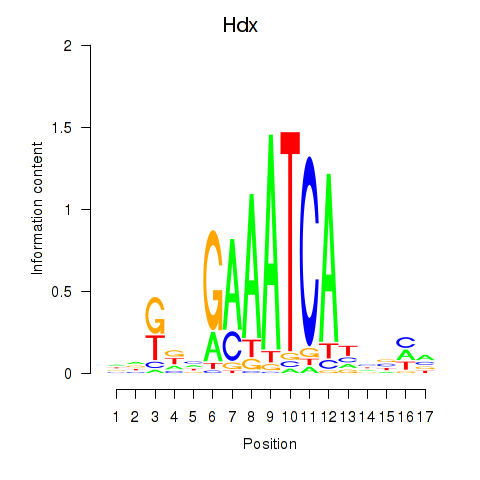
Transcription factors associated with Hdx:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hdx | ENSMUSG00000034551.6 | Hdx |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | mm10_v2_chrX_-_111697069_111697127 | 0.06 | 6.3e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 40.7 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 2.8 | 8.5 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 2.5 | 10.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 2.0 | 5.9 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 1.6 | 12.8 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 1.5 | 3.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 1.0 | 5.1 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.7 | 3.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.7 | 3.4 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.7 | 2.7 | GO:0001692 | histamine metabolic process(GO:0001692) histidine metabolic process(GO:0006547) imidazole-containing compound catabolic process(GO:0052805) |
| 0.5 | 1.5 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.5 | 9.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 6.1 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.4 | 1.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.3 | 5270.4 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5341.7 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 1.1 | 5.7 | GO:0070330 | aromatase activity(GO:0070330) |
| 1.0 | 5.9 | GO:0050786 | arachidonic acid binding(GO:0050544) RAGE receptor binding(GO:0050786) |
| 0.6 | 3.0 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.5 | 9.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.3 | 10.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.3 | 5.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.3 | 5357.8 | GO:0003674 | molecular_function(GO:0003674) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.9 | PID_TOLL_ENDOGENOUS_PATHWAY | Endogenous TLR signaling |
| 0.2 | 10.1 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.1 | 1.5 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 8.5 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 12.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID_P75_NTR_PATHWAY | p75(NTR)-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 10.1 | REACTOME_PROLACTIN_RECEPTOR_SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.4 | 9.8 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.4 | 5.1 | REACTOME_SYNTHESIS_SECRETION_AND_INACTIVATION_OF_GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.2 | 5.3 | REACTOME_HS_GAG_DEGRADATION | Genes involved in HS-GAG degradation |
| 0.2 | 3.0 | REACTOME_REGULATION_OF_INSULIN_SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.1 | 3.0 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.0 | REACTOME_ENOS_ACTIVATION_AND_REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 1.5 | REACTOME_IL1_SIGNALING | Genes involved in Interleukin-1 signaling |