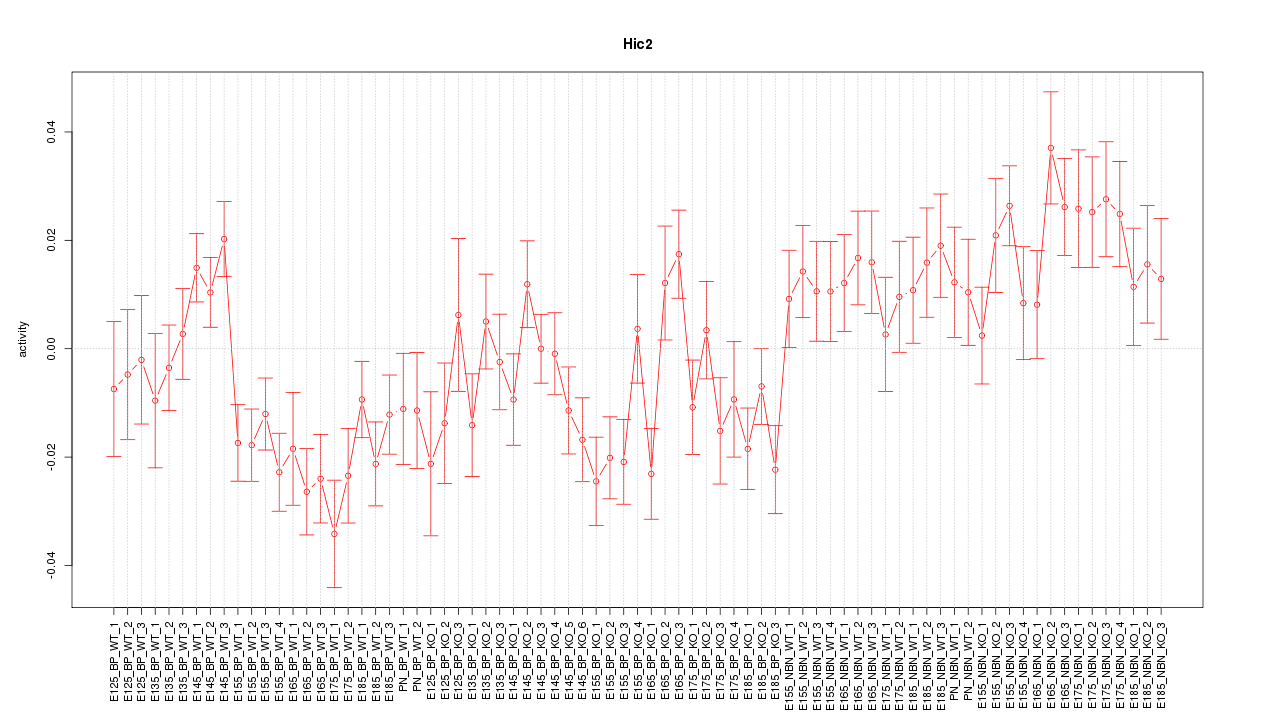
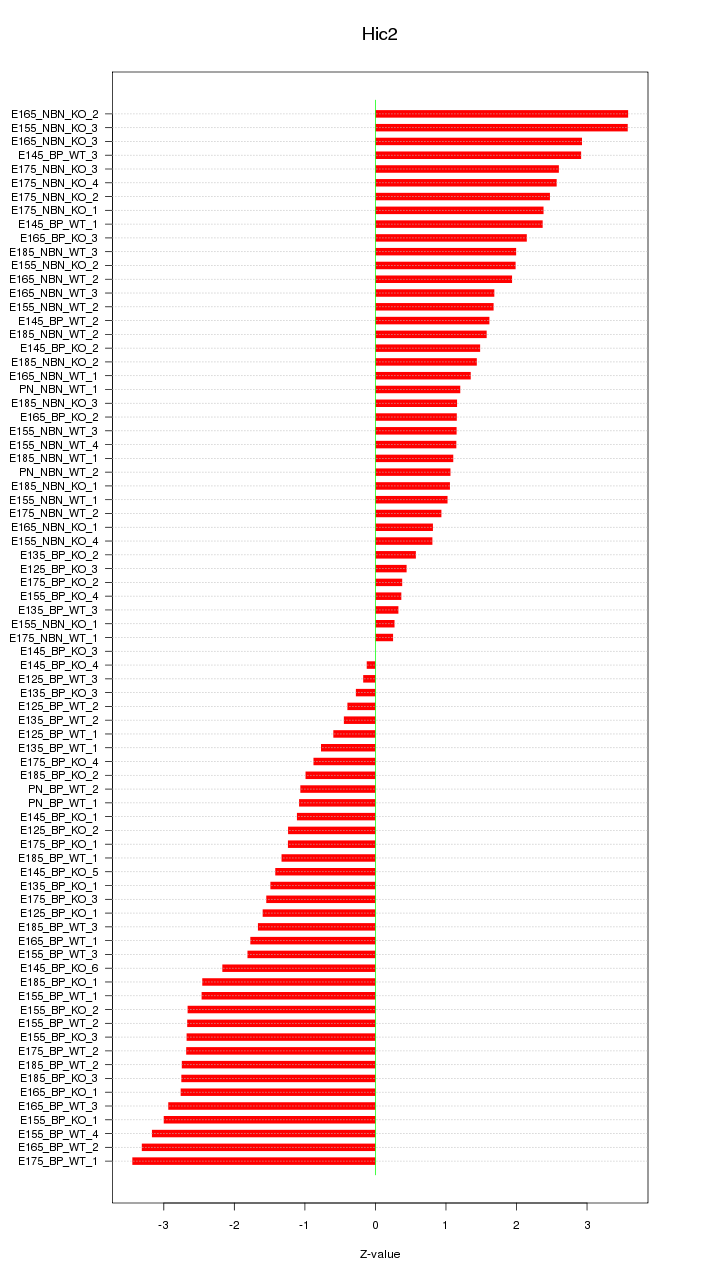
Motif ID: Hic2
Z-value: 1.871

Transcription factors associated with Hic2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hic2 | ENSMUSG00000050240.8 | Hic2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic2 | mm10_v2_chr16_+_17233560_17233664 | -0.26 | 2.2e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.5 | 19.6 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 6.4 | 57.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 3.8 | 11.3 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 3.5 | 14.2 | GO:0021586 | pons maturation(GO:0021586) |
| 3.3 | 10.0 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 3.1 | 12.3 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 2.8 | 8.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 2.8 | 8.3 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 2.7 | 10.7 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 2.6 | 10.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 2.5 | 7.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 2.1 | 12.7 | GO:0032796 | uropod organization(GO:0032796) |
| 2.1 | 10.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 2.1 | 6.3 | GO:0010752 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) |
| 2.1 | 8.4 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 2.1 | 6.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 2.1 | 12.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 2.0 | 26.6 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 2.0 | 10.0 | GO:1903609 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 2.0 | 8.0 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 2.0 | 13.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 2.0 | 5.9 | GO:0061075 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) positive regulation of amacrine cell differentiation(GO:1902871) |
| 1.9 | 31.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.9 | 1.9 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 1.9 | 5.7 | GO:0061056 | sclerotome development(GO:0061056) |
| 1.9 | 13.4 | GO:0098909 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 1.8 | 9.0 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 1.8 | 5.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 1.8 | 12.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 1.7 | 5.1 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 1.7 | 5.0 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 1.7 | 13.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 1.7 | 3.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 1.7 | 5.0 | GO:0098923 | retrograde trans-synaptic signaling(GO:0098917) retrograde trans-synaptic signaling by soluble gas(GO:0098923) |
| 1.6 | 12.9 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.6 | 7.9 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 1.5 | 6.1 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 1.5 | 4.5 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 1.5 | 3.0 | GO:0031620 | fever generation(GO:0001660) regulation of fever generation(GO:0031620) |
| 1.5 | 6.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) bud outgrowth involved in lung branching(GO:0060447) |
| 1.5 | 28.5 | GO:0001553 | luteinization(GO:0001553) |
| 1.5 | 1.5 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.4 | 2.9 | GO:0001757 | somite specification(GO:0001757) |
| 1.4 | 4.3 | GO:0009153 | purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 1.4 | 4.2 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.4 | 1.4 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 1.4 | 6.9 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 1.4 | 5.5 | GO:0099566 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 1.3 | 5.4 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 1.3 | 7.8 | GO:0097491 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.3 | 5.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.3 | 6.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.2 | 5.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 1.2 | 1.2 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 1.2 | 4.9 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.2 | 4.8 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.2 | 4.7 | GO:2000974 | negative regulation of mechanoreceptor differentiation(GO:0045632) negative regulation of pro-B cell differentiation(GO:2000974) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 1.1 | 3.4 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 1.1 | 3.4 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) |
| 1.1 | 3.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 1.1 | 16.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 1.1 | 4.3 | GO:0032275 | luteinizing hormone secretion(GO:0032275) positive regulation of gonadotropin secretion(GO:0032278) |
| 1.1 | 6.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 1.1 | 3.2 | GO:0015886 | heme transport(GO:0015886) |
| 1.1 | 4.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 1.1 | 5.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 1.0 | 2.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 1.0 | 3.1 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 1.0 | 4.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 1.0 | 11.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 1.0 | 5.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 1.0 | 3.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 1.0 | 5.0 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 1.0 | 7.0 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 1.0 | 4.8 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 1.0 | 4.8 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.9 | 14.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.9 | 2.8 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 0.9 | 2.8 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.9 | 12.7 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.9 | 1.8 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.9 | 3.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.9 | 5.2 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.9 | 2.6 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.9 | 5.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.8 | 4.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.8 | 7.6 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.8 | 2.5 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.8 | 3.3 | GO:0061743 | motor learning(GO:0061743) |
| 0.8 | 3.3 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.8 | 1.6 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.8 | 4.7 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.8 | 2.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.8 | 4.7 | GO:0046645 | positive regulation of gamma-delta T cell differentiation(GO:0045588) positive regulation of gamma-delta T cell activation(GO:0046645) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.8 | 3.9 | GO:0043084 | penile erection(GO:0043084) |
| 0.8 | 2.3 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.7 | 5.2 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.7 | 1.5 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.7 | 19.8 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.7 | 8.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.7 | 2.1 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.7 | 2.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.7 | 5.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.7 | 3.5 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.7 | 5.6 | GO:0035635 | glycoside catabolic process(GO:0016139) entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.7 | 2.7 | GO:2000277 | positive regulation of oxidative phosphorylation uncoupler activity(GO:2000277) |
| 0.7 | 2.0 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.7 | 2.0 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.7 | 3.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) T cell extravasation(GO:0072683) |
| 0.7 | 1.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.6 | 1.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.6 | 3.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.6 | 3.8 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.6 | 7.7 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.6 | 1.9 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.6 | 3.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.6 | 1.9 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.6 | 1.3 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.6 | 15.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.6 | 3.8 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) inosine biosynthetic process(GO:0046103) |
| 0.6 | 1.9 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.6 | 2.5 | GO:0033762 | taurine metabolic process(GO:0019530) response to glucagon(GO:0033762) |
| 0.6 | 1.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.6 | 3.0 | GO:0097501 | stress response to metal ion(GO:0097501) |
| 0.6 | 18.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.6 | 6.3 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.6 | 1.7 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.6 | 7.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.6 | 2.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.5 | 11.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.5 | 1.1 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.5 | 0.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.5 | 3.2 | GO:0090343 | positive regulation of cell aging(GO:0090343) |
| 0.5 | 6.4 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.5 | 1.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.5 | 3.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.5 | 5.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.5 | 2.6 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.5 | 1.5 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K6-linked deubiquitination(GO:0044313) |
| 0.5 | 2.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.5 | 1.5 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.5 | 2.0 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.5 | 7.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.5 | 5.1 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.5 | 6.5 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 2.0 | GO:0002924 | negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.5 | 1.5 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.5 | 1.5 | GO:0071544 | diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.5 | 1.5 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 0.5 | 2.0 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.5 | 4.9 | GO:0061470 | interleukin-21 production(GO:0032625) T follicular helper cell differentiation(GO:0061470) interleukin-21 secretion(GO:0072619) |
| 0.5 | 11.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.5 | 1.9 | GO:1903898 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.5 | 4.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.5 | 0.9 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.5 | 2.4 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.5 | 1.4 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) connective tissue replacement(GO:0097709) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) regulation of connective tissue replacement(GO:1905203) |
| 0.5 | 1.9 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.5 | 3.7 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.5 | 1.4 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.5 | 2.3 | GO:1901731 | positive regulation of platelet aggregation(GO:1901731) |
| 0.5 | 5.9 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 2.3 | GO:0002016 | regulation of blood volume by renin-angiotensin(GO:0002016) |
| 0.4 | 3.1 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.4 | 3.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.4 | 5.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.4 | 2.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.4 | 6.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.4 | 7.7 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.4 | 1.3 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.4 | 1.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.4 | 5.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.4 | 2.1 | GO:0045346 | regulation of MHC class II biosynthetic process(GO:0045346) negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.4 | 0.8 | GO:0070970 | interleukin-2 secretion(GO:0070970) |
| 0.4 | 2.0 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.4 | 1.6 | GO:0002339 | B cell selection(GO:0002339) |
| 0.4 | 2.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.4 | 2.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 2.0 | GO:1903772 | regulation of viral budding via host ESCRT complex(GO:1903772) |
| 0.4 | 2.4 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.4 | 3.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) lipid glycosylation(GO:0030259) |
| 0.4 | 3.9 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.4 | 6.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 4.2 | GO:1990173 | protein localization to nuclear body(GO:1903405) positive regulation of establishment of protein localization to telomere(GO:1904851) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.4 | 3.0 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 3.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.4 | 9.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.4 | 1.1 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) formin-nucleated actin cable assembly(GO:0070649) |
| 0.3 | 1.0 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 1.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.3 | 5.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.3 | 11.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.3 | 7.3 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.3 | 2.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 1.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.3 | 3.2 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.3 | 3.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 2.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.3 | 1.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.3 | 2.5 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.3 | 7.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 2.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.3 | 1.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.3 | 1.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 | 4.0 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.3 | 1.5 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.3 | 1.2 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.3 | 0.6 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 3.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.3 | 3.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.3 | 3.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.3 | 10.9 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.3 | 2.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.3 | 2.0 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 2.0 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 2.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.3 | 2.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.3 | 13.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.3 | 1.4 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.3 | 3.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.3 | 5.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.3 | 0.3 | GO:0016137 | glycoside metabolic process(GO:0016137) |
| 0.3 | 1.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.3 | 4.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 0.5 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.3 | 1.3 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.3 | 4.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.3 | 1.6 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.3 | 0.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 2.8 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.3 | 3.3 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.2 | 0.7 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 6.7 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 | 6.9 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.2 | 2.2 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.2 | 3.0 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 3.9 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.2 | 6.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 0.5 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.2 | 0.7 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.2 | 10.8 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.2 | 1.1 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.2 | 2.0 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 2.4 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.2 | 0.9 | GO:0061450 | trophoblast cell migration(GO:0061450) regulation of trophoblast cell migration(GO:1901163) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.2 | 2.4 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.2 | 0.9 | GO:0001907 | killing by symbiont of host cells(GO:0001907) induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) disruption by symbiont of host cell(GO:0044004) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.2 | 1.3 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.2 | 2.5 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.2 | 1.0 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.2 | 0.8 | GO:0019732 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.2 | 1.2 | GO:0045954 | positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.2 | 0.6 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 0.8 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.2 | 1.0 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.2 | 1.8 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.4 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 3.1 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.2 | 1.4 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.2 | 7.1 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.2 | 3.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.2 | 3.0 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.2 | 1.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.2 | 1.9 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.2 | 0.7 | GO:0032532 | regulation of microvillus length(GO:0032532) |
| 0.2 | 0.4 | GO:0006550 | isoleucine metabolic process(GO:0006549) isoleucine catabolic process(GO:0006550) |
| 0.2 | 4.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 2.0 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.2 | 0.7 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 1.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 11.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 1.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.2 | 0.3 | GO:0043379 | memory T cell differentiation(GO:0043379) |
| 0.2 | 0.7 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.2 | 1.5 | GO:2001197 | positive regulation of extracellular matrix assembly(GO:1901203) regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.2 | 2.2 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 0.5 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.2 | 3.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.2 | 0.5 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.2 | 0.5 | GO:2001013 | adherens junction maintenance(GO:0034334) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.2 | 0.6 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.2 | 0.6 | GO:1903319 | positive regulation of protein maturation(GO:1903319) |
| 0.2 | 0.6 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.2 | 0.6 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.2 | 1.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.2 | 3.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.2 | 2.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.1 | 1.8 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 2.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.9 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 2.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.3 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.1 | 0.7 | GO:0046479 | glycolipid catabolic process(GO:0019377) glycosphingolipid catabolic process(GO:0046479) |
| 0.1 | 1.5 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 2.4 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.5 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 1.7 | GO:0048820 | hair follicle maturation(GO:0048820) |
| 0.1 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 8.5 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.1 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.7 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 2.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.1 | 3.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 3.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.3 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 6.8 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 1.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.5 | GO:0015817 | histidine transport(GO:0015817) |
| 0.1 | 0.3 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 1.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.9 | GO:0032309 | icosanoid secretion(GO:0032309) |
| 0.1 | 0.3 | GO:0010455 | positive regulation of cell fate commitment(GO:0010455) |
| 0.1 | 0.7 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 5.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.1 | GO:0044333 | planar cell polarity pathway involved in axis elongation(GO:0003402) Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.1 | 2.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 1.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 3.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 2.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.1 | 1.8 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.8 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.9 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.1 | 1.7 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.1 | 0.6 | GO:1903056 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.1 | 1.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 4.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.4 | GO:0050716 | regulation of interleukin-1 secretion(GO:0050704) regulation of interleukin-1 beta secretion(GO:0050706) positive regulation of interleukin-1 secretion(GO:0050716) positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 0.4 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 3.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.6 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 2.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 2.4 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.1 | 1.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 0.4 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 4.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 1.2 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.1 | GO:0015810 | aspartate transport(GO:0015810) |
| 0.1 | 2.3 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 2.8 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 1.1 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 0.6 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 1.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.2 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.1 | 3.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 1.5 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.1 | 1.8 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 1.6 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 2.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 6.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 1.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.2 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 4.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 0.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 3.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.8 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.1 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 1.0 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.1 | GO:0051126 | negative regulation of actin nucleation(GO:0051126) |
| 0.0 | 0.3 | GO:0006497 | protein lipidation(GO:0006497) lipoprotein biosynthetic process(GO:0042158) |
| 0.0 | 2.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 2.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.5 | GO:0015800 | acidic amino acid transport(GO:0015800) |
| 0.0 | 2.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.9 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 2.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.8 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.4 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 1.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.1 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.0 | 1.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.8 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 2.4 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 1.4 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 1.3 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 1.1 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0032438 | melanosome organization(GO:0032438) pigment granule organization(GO:0048753) |
| 0.0 | 0.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.2 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 1.0 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.3 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.3 | GO:1901016 | regulation of potassium ion transmembrane transporter activity(GO:1901016) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.7 | 57.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 4.7 | 14.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 3.5 | 10.5 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 3.1 | 12.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 2.2 | 8.9 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 2.0 | 2.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 1.9 | 11.7 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 1.9 | 9.5 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 1.8 | 5.4 | GO:0098855 | HCN channel complex(GO:0098855) |
| 1.7 | 10.4 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 1.6 | 8.0 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 1.5 | 30.2 | GO:0098984 | neuron to neuron synapse(GO:0098984) |
| 1.1 | 3.3 | GO:0044299 | C-fiber(GO:0044299) |
| 1.1 | 17.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 1.1 | 3.2 | GO:0044194 | cytolytic granule(GO:0044194) |
| 1.1 | 11.7 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.1 | 5.3 | GO:0032437 | cuticular plate(GO:0032437) |
| 1.0 | 3.0 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 1.0 | 15.8 | GO:0043196 | varicosity(GO:0043196) |
| 0.9 | 11.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.9 | 14.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.9 | 14.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.8 | 3.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.8 | 7.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.8 | 10.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.8 | 10.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.7 | 6.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.7 | 2.2 | GO:0098835 | presynaptic endosome(GO:0098830) presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.7 | 33.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.7 | 2.1 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.7 | 4.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 2.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.7 | 3.4 | GO:1990357 | terminal web(GO:1990357) |
| 0.7 | 41.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.6 | 2.6 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.6 | 9.0 | GO:0016589 | NURF complex(GO:0016589) |
| 0.6 | 18.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.6 | 2.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.6 | 12.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.6 | 24.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.6 | 2.9 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 4.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.6 | 2.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.5 | 1.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.5 | 6.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.5 | 1.6 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.5 | 1.5 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 7.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.5 | 2.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.5 | 1.5 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 5.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.5 | 1.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.5 | 3.8 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.5 | 6.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.5 | 2.3 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 47.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.5 | 3.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 14.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 1.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 0.9 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.4 | 1.6 | GO:0044301 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) |
| 0.4 | 4.9 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.4 | 1.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 1.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.6 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.4 | 2.4 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.4 | 4.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.4 | 2.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.4 | 4.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.4 | 5.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.4 | 1.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.4 | 9.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.4 | 7.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.3 | 0.7 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 0.3 | 1.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.3 | 4.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.3 | 2.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.3 | 2.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.3 | 1.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.3 | 1.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.3 | 1.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.3 | 6.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.3 | 3.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.3 | 4.6 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.3 | 2.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.3 | 2.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 4.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.3 | 2.8 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.3 | 1.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.3 | 3.0 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 6.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 10.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.2 | 4.7 | GO:0038201 | TOR complex(GO:0038201) |
| 0.2 | 6.7 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 8.4 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.2 | 5.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.2 | 25.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.2 | 1.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.2 | 1.5 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.2 | 0.6 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.2 | 8.2 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 1.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 1.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.2 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 0.9 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.2 | 0.7 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.2 | 1.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.2 | 1.4 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.2 | 3.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 2.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 2.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 2.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 1.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.8 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 0.9 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.9 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 1.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 5.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 15.9 | GO:0044309 | neuron spine(GO:0044309) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.7 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 1.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 5.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 1.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 4.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.1 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 2.6 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 1.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.1 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 13.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 2.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.7 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 1.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 3.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 4.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.1 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 7.0 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 2.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 4.3 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 2.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 9.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 2.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.7 | GO:0022627 | small ribosomal subunit(GO:0015935) cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 2.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 3.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 1.0 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.9 | 55.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 3.9 | 27.6 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 3.6 | 14.3 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 3.3 | 10.0 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 3.1 | 12.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.8 | 19.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 2.6 | 13.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 2.3 | 6.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.2 | 8.9 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 2.2 | 6.6 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.1 | 10.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 2.1 | 8.4 | GO:0070573 | tripeptidyl-peptidase activity(GO:0008240) metallodipeptidase activity(GO:0070573) |
| 2.1 | 12.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 2.0 | 10.0 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 1.9 | 5.6 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 1.8 | 5.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 1.6 | 9.6 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 1.6 | 14.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.5 | 4.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 1.4 | 4.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.4 | 6.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 1.4 | 5.5 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 1.3 | 5.4 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.3 | 5.2 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.3 | 5.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.2 | 3.7 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 1.2 | 12.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 1.1 | 2.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 1.1 | 4.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 1.1 | 2.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.1 | 3.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.1 | 17.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 1.1 | 3.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 1.1 | 3.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 1.0 | 5.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.0 | 5.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 1.0 | 6.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 1.0 | 13.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.0 | 2.9 | GO:0030519 | snoRNP binding(GO:0030519) |
| 1.0 | 3.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.9 | 12.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.9 | 6.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.9 | 7.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.9 | 2.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.9 | 5.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.9 | 9.5 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.9 | 2.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.8 | 8.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.8 | 3.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.8 | 3.3 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.8 | 9.9 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.8 | 2.3 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.8 | 2.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.8 | 5.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.8 | 4.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.8 | 3.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.7 | 14.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.7 | 25.9 | GO:0022839 | ion gated channel activity(GO:0022839) |
| 0.7 | 2.1 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.7 | 5.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.7 | 16.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.7 | 2.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.6 | 1.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.6 | 1.9 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.6 | 5.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.6 | 5.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.6 | 4.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.6 | 0.6 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.6 | 2.5 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.6 | 2.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.6 | 5.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.6 | 4.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.6 | 3.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.6 | 17.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.6 | 2.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.5 | 9.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.5 | 1.6 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.5 | 11.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.5 | 3.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.5 | 4.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 3.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.5 | 1.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.5 | 1.5 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.5 | 14.0 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.5 | 9.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.5 | 1.4 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.5 | 1.9 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.5 | 1.9 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.5 | 1.4 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.5 | 2.8 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.5 | 1.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.4 | 7.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.4 | 3.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.4 | 3.9 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.4 | 1.7 | GO:0043546 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.4 | 10.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.4 | 3.0 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.4 | 2.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.4 | 3.8 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.4 | 3.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.4 | 15.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.4 | 2.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 3.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.4 | 1.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.4 | 2.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.4 | 1.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.4 | 4.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.5 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.4 | 1.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 3.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.4 | 2.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.4 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.4 | 1.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.4 | 1.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.4 | 15.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 2.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.3 | 3.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.3 | 1.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.3 | 3.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.3 | 3.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.3 | 10.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.3 | 2.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.3 | 3.6 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.3 | 1.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 5.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 2.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.3 | 1.5 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.3 | 1.2 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 1.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.3 | 5.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 2.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 2.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.3 | 2.4 | GO:0070290 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 1.2 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.3 | 2.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 3.8 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.3 | 1.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 2.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.3 | 0.9 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.3 | 5.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.3 | 1.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.3 | 1.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.3 | 1.6 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.3 | 1.9 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.3 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 8.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.2 | 0.7 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 1.5 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 1.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 3.7 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 3.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 5.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.2 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.2 | 1.9 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 4.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.2 | 1.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 3.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.2 | 2.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 2.3 | GO:0044213 | intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.2 | 6.0 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.0 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.2 | 2.5 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 5.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 0.6 | GO:0098973 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 6.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 2.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.2 | 0.6 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.2 | 1.8 | GO:0032564 | dATP binding(GO:0032564) |
| 0.2 | 9.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.2 | 5.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 1.5 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.2 | 4.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 2.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.2 | 1.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 8.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 1.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.2 | 0.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.2 | 0.5 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.2 | 1.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.1 | 0.7 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 2.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 1.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.5 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.1 | 2.8 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 11.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 2.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 3.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.3 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 2.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.3 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 3.5 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 1.6 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.4 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.1 | 10.0 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 2.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 2.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.6 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 4.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 5.9 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 2.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 2.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.6 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.7 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.9 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.7 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 3.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 7.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 2.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 5.2 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 2.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 2.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.9 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 4.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 10.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 8.6 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.4 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.6 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.5 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.3 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 1.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 6.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.1 | 1.8 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 2.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.2 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.1 | 0.4 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 1.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 1.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 4.9 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 3.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 3.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 2.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 2.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 57.4 | PID_P38_GAMMA_DELTA_PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.0 | 15.1 | PID_ERBB_NETWORK_PATHWAY | ErbB receptor signaling network |
| 0.8 | 8.4 | SA_MMP_CYTOKINE_CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.7 | 2.2 | PID_REELIN_PATHWAY | Reelin signaling pathway |
| 0.4 | 3.5 | PID_VEGF_VEGFR_PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 12.1 | PID_INTEGRIN_A9B1_PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 3.4 | PID_S1P_S1P4_PATHWAY | S1P4 pathway |
| 0.4 | 11.5 | PID_ARF_3PATHWAY | Arf1 pathway |
| 0.4 | 14.1 | PID_RXR_VDR_PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.4 | 9.7 | NABA_BASEMENT_MEMBRANES | Genes encoding structural components of basement membranes |
| 0.4 | 7.1 | ST_G_ALPHA_S_PATHWAY | G alpha s Pathway |
| 0.4 | 17.7 | SIG_CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.3 | 8.7 | PID_KIT_PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.3 | 14.2 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 14.1 | PID_AMB2_NEUTROPHILS_PATHWAY | amb2 Integrin signaling |
| 0.3 | 12.7 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.3 | 13.4 | ST_T_CELL_SIGNAL_TRANSDUCTION | T Cell Signal Transduction |
| 0.3 | 16.9 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 12.3 | PID_BMP_PATHWAY | BMP receptor signaling |
| 0.3 | 5.4 | SA_PTEN_PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.3 | 1.8 | PID_RET_PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.3 | 4.9 | PID_CIRCADIAN_PATHWAY | Circadian rhythm pathway |
| 0.2 | 2.2 | PID_UPA_UPAR_PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.2 | 3.3 | PID_IL23_PATHWAY | IL23-mediated signaling events |
| 0.2 | 8.0 | PID_TCPTP_PATHWAY | Signaling events mediated by TCPTP |
| 0.2 | 5.4 | PID_FGF_PATHWAY | FGF signaling pathway |
| 0.2 | 7.1 | PID_INTEGRIN3_PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 9.1 | PID_P38_ALPHA_BETA_DOWNSTREAM_PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 21.5 | PID_PDGFRB_PATHWAY | PDGFR-beta signaling pathway |
| 0.2 | 2.4 | PID_ANGIOPOIETIN_RECEPTOR_PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.2 | 4.2 | PID_FRA_PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 5.9 | PID_HNF3B_PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.2 | 5.4 | PID_ATF2_PATHWAY | ATF-2 transcription factor network |
| 0.2 | 3.8 | PID_FAK_PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 10.3 | PID_MYC_REPRESS_PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 5.0 | NABA_PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 22.4 | NABA_SECRETED_FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.0 | PID_IL3_PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.3 | SA_REG_CASCADE_OF_CYCLIN_EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 7.4 | PID_NOTCH_PATHWAY | Notch signaling pathway |
| 0.1 | 1.6 | PID_RAC1_PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.0 | PID_LKB1_PATHWAY | LKB1 signaling events |
| 0.1 | 3.1 | PID_ARF6_TRAFFICKING_PATHWAY | Arf6 trafficking events |
| 0.1 | 0.7 | PID_IL4_2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.5 | PID_HIF1A_PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.5 | PID_RB_1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.1 | ST_INTEGRIN_SIGNALING_PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.9 | PID_HIF1_TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 1.8 | PID_AP1_PATHWAY | AP-1 transcription factor network |
| 0.1 | 3.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 1.9 | PID_DELTA_NP63_PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.0 | SIG_REGULATION_OF_THE_ACTIN_CYTOSKELETON_BY_RHO_GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.4 | ST_TYPE_I_INTERFERON_PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.2 | PID_SYNDECAN_4_PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.0 | PID_RHOA_PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID_ATM_PATHWAY | ATM pathway |
| 0.0 | 2.1 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID_EPHA2_FWD_PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.8 | PID_BETA_CATENIN_DEG_PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | SIG_CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID_AURORA_A_PATHWAY | Aurora A signaling |
| 0.0 | 0.1 | PID_NECTIN_PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.5 | NABA_ECM_REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID_MYC_PATHWAY | C-MYC pathway |
| 0.0 | 0.3 | PID_RAS_PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID_EPHA_FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 26.2 | REACTOME_TRANSPORT_OF_ORGANIC_ANIONS | Genes involved in Transport of organic anions |
| 2.5 | 56.7 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 1.9 | 24.2 | REACTOME_NUCLEOTIDE_LIKE_PURINERGIC_RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 1.4 | 14.3 | REACTOME_ACETYLCHOLINE_NEUROTRANSMITTER_RELEASE_CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 1.4 | 9.5 | REACTOME_TANDEM_PORE_DOMAIN_POTASSIUM_CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.1 | 24.2 | REACTOME_HORMONE_SENSITIVE_LIPASE_HSL_MEDIATED_TRIACYLGLYCEROL_HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.8 | 10.6 | REACTOME_AMINE_LIGAND_BINDING_RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.6 | 7.7 | REACTOME_SIGNAL_REGULATORY_PROTEIN_SIRP_FAMILY_INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.6 | 16.0 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 7.6 | REACTOME_GABA_SYNTHESIS_RELEASE_REUPTAKE_AND_DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.6 | 8.8 | REACTOME_ACTIVATION_OF_RAC | Genes involved in Activation of Rac |
| 0.5 | 6.5 | REACTOME_CD28_DEPENDENT_VAV1_PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.5 | 4.3 | REACTOME_FGFR4_LIGAND_BINDING_AND_ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 14.5 | REACTOME_SIGNALING_BY_BMP | Genes involved in Signaling by BMP |
| 0.4 | 7.9 | REACTOME_SHC1_EVENTS_IN_ERBB4_SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.4 | 12.2 | REACTOME_RAP1_SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 7.4 | REACTOME_PURINE_SALVAGE | Genes involved in Purine salvage |
| 0.4 | 5.1 | REACTOME_VEGF_LIGAND_RECEPTOR_INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 3.4 | REACTOME_NA_CL_DEPENDENT_NEUROTRANSMITTER_TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.4 | 7.6 | REACTOME_FORMATION_OF_ATP_BY_CHEMIOSMOTIC_COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.4 | 11.1 | REACTOME_RAS_ACTIVATION_UOPN_CA2_INFUX_THROUGH_NMDA_RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.4 | 7.7 | REACTOME_GAP_JUNCTION_DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 13.6 | REACTOME_TRAFFICKING_OF_AMPA_RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.4 | 13.0 | REACTOME_INTERACTION_BETWEEN_L1_AND_ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.4 | 5.1 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.3 | 17.1 | REACTOME_NCAM1_INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 2.6 | REACTOME_APOBEC3G_MEDIATED_RESISTANCE_TO_HIV1_INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.3 | 4.2 | REACTOME_CLASS_C_3_METABOTROPIC_GLUTAMATE_PHEROMONE_RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 1.6 | REACTOME_NCAM_SIGNALING_FOR_NEURITE_OUT_GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.3 | 2.7 | REACTOME_ACTIVATION_OF_CHAPERONE_GENES_BY_ATF6_ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 23.4 | REACTOME_PEPTIDE_LIGAND_BINDING_RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 7.9 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 11.8 | REACTOME_GLYCOLYSIS | Genes involved in Glycolysis |
| 0.3 | 17.6 | REACTOME_NUCLEAR_RECEPTOR_TRANSCRIPTION_PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.3 | 2.3 | REACTOME_GAMMA_CARBOXYLATION_TRANSPORT_AND_AMINO_TERMINAL_CLEAVAGE_OF_PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 4.3 | REACTOME_ANTIGEN_PRESENTATION_FOLDING_ASSEMBLY_AND_PEPTIDE_LOADING_OF_CLASS_I_MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.3 | 2.5 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PG | Genes involved in Acyl chain remodelling of PG |
| 0.3 | 2.3 | REACTOME_RECRUITMENT_OF_NUMA_TO_MITOTIC_CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.3 | 4.1 | REACTOME_PRE_NOTCH_PROCESSING_IN_GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 2.0 | REACTOME_KERATAN_SULFATE_DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 7.8 | REACTOME_O_LINKED_GLYCOSYLATION_OF_MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.2 | 4.2 | REACTOME_FORMATION_OF_TUBULIN_FOLDING_INTERMEDIATES_BY_CCT_TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 7.3 | REACTOME_ACTIVATED_NOTCH1_TRANSMITS_SIGNAL_TO_THE_NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 6.2 | REACTOME_G_PROTEIN_ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 4.5 | REACTOME_POST_NMDA_RECEPTOR_ACTIVATION_EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.2 | 3.0 | REACTOME_ZINC_TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 3.8 | REACTOME_BASIGIN_INTERACTIONS | Genes involved in Basigin interactions |
| 0.2 | 5.2 | REACTOME_A_TETRASACCHARIDE_LINKER_SEQUENCE_IS_REQUIRED_FOR_GAG_SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.2 | 2.8 | REACTOME_PTM_GAMMA_CARBOXYLATION_HYPUSINE_FORMATION_AND_ARYLSULFATASE_ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 3.4 | REACTOME_EFFECTS_OF_PIP2_HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.2 | 2.5 | REACTOME_SYNTHESIS_SECRETION_AND_DEACYLATION_OF_GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.9 | REACTOME_NEPHRIN_INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 1.5 | REACTOME_GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.8 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.1 | REACTOME_PLATELET_CALCIUM_HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.7 | REACTOME_ROLE_OF_DCC_IN_REGULATING_APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 2.5 | REACTOME_SULFUR_AMINO_ACID_METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.5 | REACTOME_INSULIN_SYNTHESIS_AND_PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 1.8 | REACTOME_SIGNALING_BY_NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 8.9 | REACTOME_POTASSIUM_CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 2.1 | REACTOME_METABOLISM_OF_POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.0 | REACTOME_HS_GAG_BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 1.6 | REACTOME_CYTOCHROME_P450_ARRANGED_BY_SUBSTRATE_TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.4 | REACTOME_GPVI_MEDIATED_ACTIVATION_CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.8 | REACTOME_TRANSPORT_TO_THE_GOLGI_AND_SUBSEQUENT_MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.3 | REACTOME_AMINE_COMPOUND_SLC_TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 3.0 | REACTOME_CHOLESTEROL_BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.4 | REACTOME_CYTOSOLIC_TRNA_AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.2 | REACTOME_DESTABILIZATION_OF_MRNA_BY_AUF1_HNRNP_D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 5.8 | REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 3.7 | REACTOME_BMAL1_CLOCK_NPAS2_ACTIVATES_CIRCADIAN_EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.4 | REACTOME_POL_SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.3 | REACTOME_CHONDROITIN_SULFATE_BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 1.8 | REACTOME_PROCESSING_OF_INTRONLESS_PRE_MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 3.0 | REACTOME_LATENT_INFECTION_OF_HOMO_SAPIENS_WITH_MYCOBACTERIUM_TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.0 | REACTOME_CALNEXIN_CALRETICULIN_CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.4 | REACTOME_HDL_MEDIATED_LIPID_TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 2.2 | REACTOME_CIRCADIAN_CLOCK | Genes involved in Circadian Clock |
| 0.1 | 1.5 | REACTOME_OXYGEN_DEPENDENT_PROLINE_HYDROXYLATION_OF_HYPOXIA_INDUCIBLE_FACTOR_ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.9 | REACTOME_SYNTHESIS_OF_PC | Genes involved in Synthesis of PC |
| 0.1 | 3.3 | REACTOME_SYNTHESIS_OF_PIPS_AT_THE_PLASMA_MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME_DOWNREGULATION_OF_TGF_BETA_RECEPTOR_SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 1.3 | REACTOME_CYCLIN_A_B1_ASSOCIATED_EVENTS_DURING_G2_M_TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME_ERKS_ARE_INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.1 | REACTOME_SIGNALING_BY_PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 2.6 | REACTOME_TRANSPORT_OF_VITAMINS_NUCLEOSIDES_AND_RELATED_MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 4.2 | REACTOME_GOLGI_ASSOCIATED_VESICLE_BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.5 | REACTOME_ROLE_OF_SECOND_MESSENGERS_IN_NETRIN1_SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 4.6 | REACTOME_RESPIRATORY_ELECTRON_TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.4 | REACTOME_TIE2_SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 1.7 | REACTOME_SMOOTH_MUSCLE_CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 4.2 | REACTOME_MEIOTIC_SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 3.1 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 2.6 | REACTOME_MITOCHONDRIAL_PROTEIN_IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.2 | REACTOME_ALPHA_LINOLENIC_ACID_ALA_METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.1 | REACTOME_REGULATION_OF_ORNITHINE_DECARBOXYLASE_ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 0.6 | REACTOME_PREFOLDIN_MEDIATED_TRANSFER_OF_SUBSTRATE_TO_CCT_TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.2 | REACTOME_TRANSPORT_OF_MATURE_MRNA_DERIVED_FROM_AN_INTRONLESS_TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.8 | REACTOME_UNWINDING_OF_DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME_CGMP_EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.4 | REACTOME_REGULATION_OF_IFNA_SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME_PASSIVE_TRANSPORT_BY_AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME_MRNA_SPLICING_MINOR_PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME_REGULATION_OF_RHEB_GTPASE_ACTIVITY_BY_AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.6 | REACTOME_GLYCOSPHINGOLIPID_METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME_ACYL_CHAIN_REMODELLING_OF_PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.2 | REACTOME_PEPTIDE_CHAIN_ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME_BRANCHED_CHAIN_AMINO_ACID_CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME_RNA_POL_III_CHAIN_ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.2 | REACTOME_TRANSPORT_OF_INORGANIC_CATIONS_ANIONS_AND_AMINO_ACIDS_OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME_TRANSCRIPTIONAL_REGULATION_OF_WHITE_ADIPOCYTE_DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |