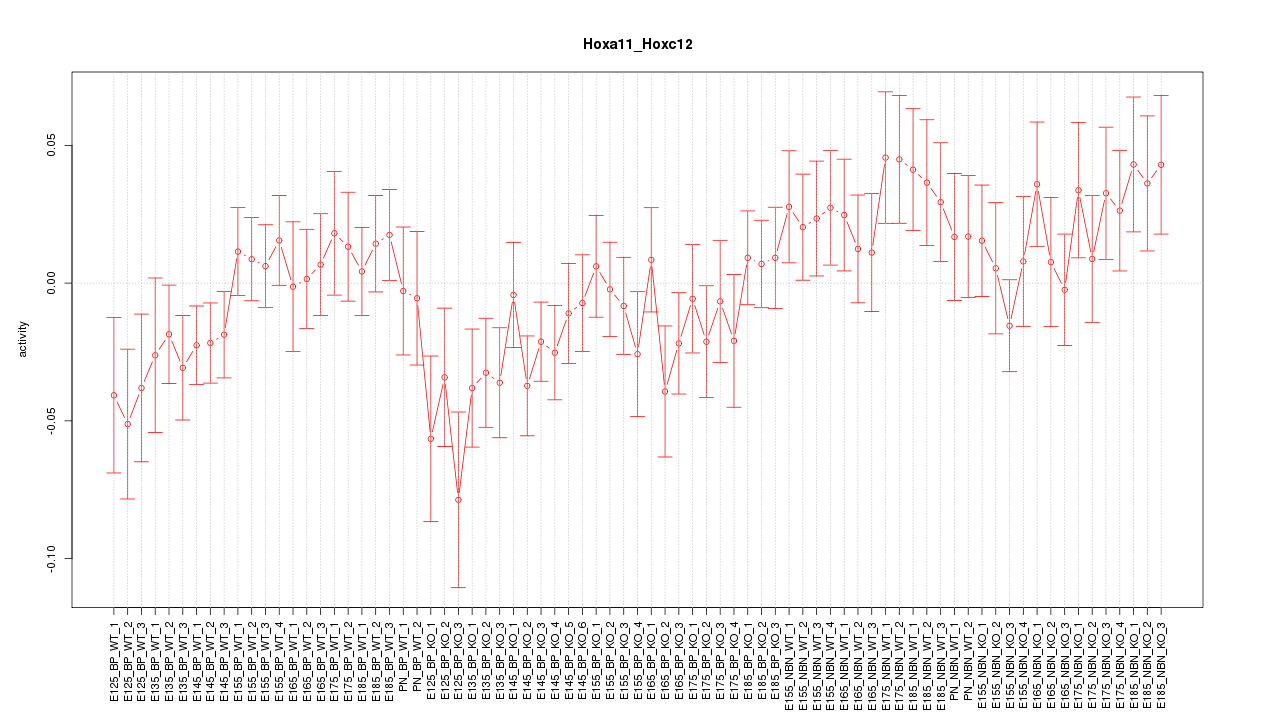
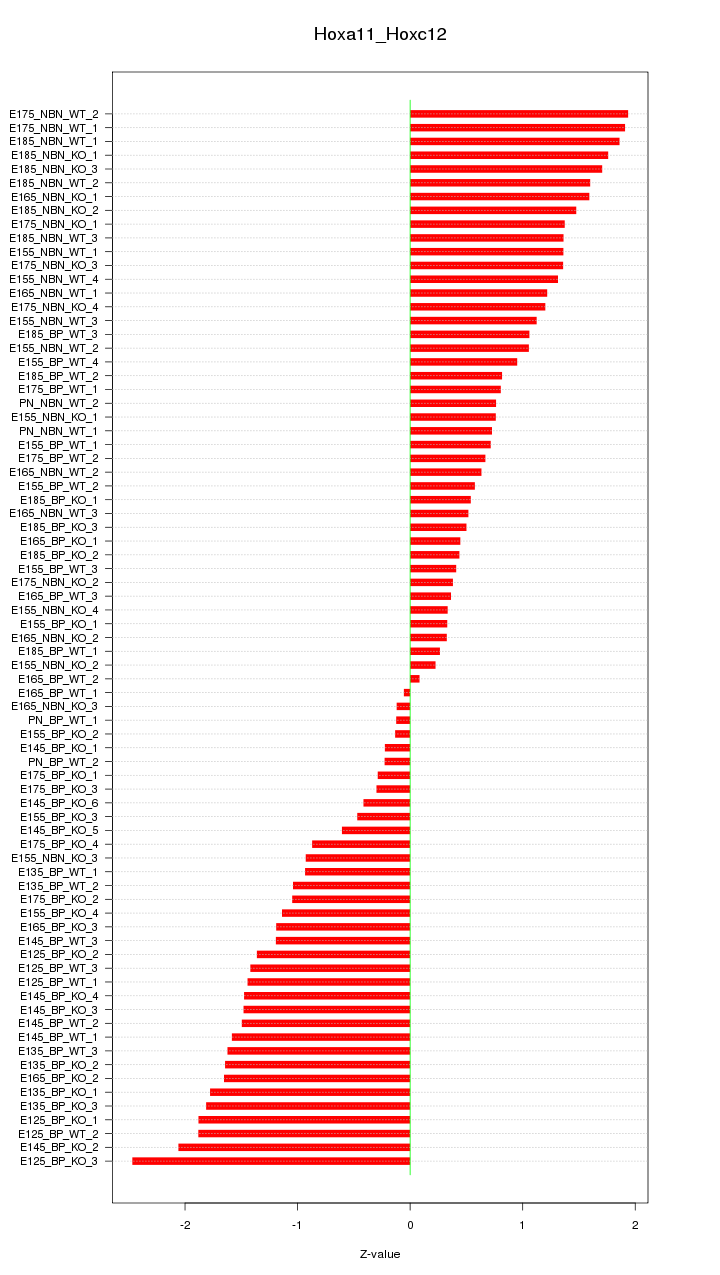
Motif ID: Hoxa11_Hoxc12
Z-value: 1.164
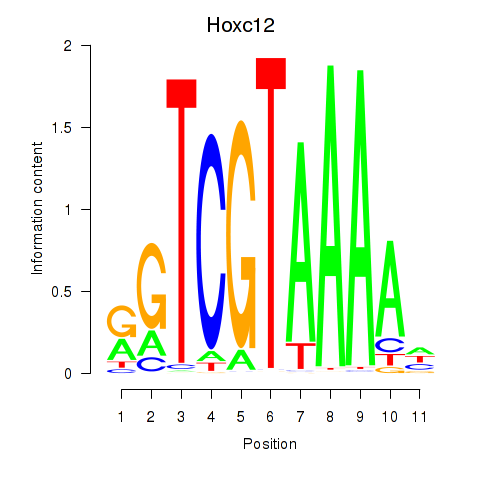
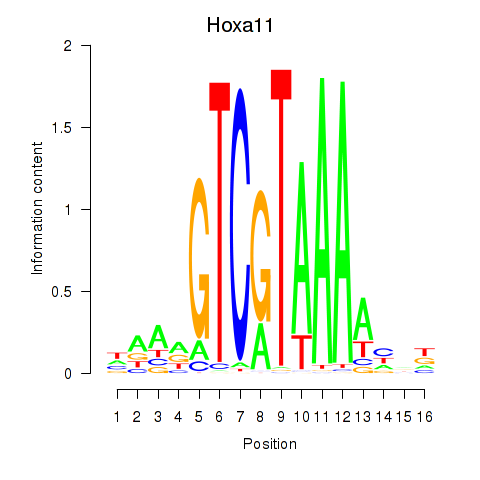
Transcription factors associated with Hoxa11_Hoxc12:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa11 | ENSMUSG00000038210.9 | Hoxa11 |
| Hoxc12 | ENSMUSG00000050328.2 | Hoxc12 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 14.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 3.8 | 18.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) progesterone secretion(GO:0042701) negative regulation of hair follicle development(GO:0051799) positive regulation of ovulation(GO:0060279) |
| 2.9 | 17.5 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 2.0 | 5.9 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 1.9 | 5.7 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 1.4 | 11.2 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.4 | 22.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.4 | 8.2 | GO:0009405 | pathogenesis(GO:0009405) |
| 1.3 | 3.8 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 1.1 | 8.6 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 1.1 | 4.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.0 | 4.0 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.9 | 6.0 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.8 | 2.5 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.7 | 2.1 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.7 | 5.3 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.7 | 19.1 | GO:0022401 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) |
| 0.7 | 2.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.5 | 2.7 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.5 | 2.6 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.4 | 1.7 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.4 | 1.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.4 | 4.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.4 | 2.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.4 | 9.7 | GO:0030903 | notochord development(GO:0030903) |
| 0.3 | 2.0 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.3 | 1.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.3 | 1.9 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 2.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 3.1 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.3 | 1.3 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.3 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 3.2 | GO:1901898 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 10.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.2 | 0.9 | GO:0015744 | succinate transport(GO:0015744) |
| 0.2 | 1.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.2 | 0.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.7 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.0 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 3.1 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 6.7 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.1 | 1.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.8 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.1 | 0.6 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.4 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 1.7 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 2.4 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 1.4 | GO:0036120 | response to platelet-derived growth factor(GO:0036119) cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 1.6 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 5.3 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 2.3 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.1 | 0.6 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 2.1 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 2.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.0 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 8.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.1 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.0 | 0.4 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.0 | 5.2 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.4 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 1.2 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 2.6 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.8 | GO:0043512 | inhibin A complex(GO:0043512) |
| 5.8 | 17.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 1.2 | 6.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.8 | 2.5 | GO:0031417 | NatC complex(GO:0031417) |
| 0.8 | 10.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.7 | 11.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.6 | 18.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.5 | 3.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.4 | 6.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.4 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 1.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 23.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 2.0 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 1.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 2.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.1 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 3.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 2.1 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 4.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 0.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 3.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 5.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 3.8 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 1.0 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.3 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.4 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 2.0 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 5.2 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.4 | 19.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 5.8 | 17.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 3.1 | 18.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 2.4 | 14.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 1.1 | 4.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.1 | 4.2 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.0 | 6.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.9 | 5.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.8 | 4.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.8 | 2.3 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.7 | 2.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.6 | 2.6 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.6 | 8.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.6 | 2.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 3.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 0.9 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.3 | 8.6 | GO:0043539 | insulin-like growth factor receptor binding(GO:0005159) protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 3.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.3 | 2.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.3 | 3.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.3 | 0.8 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.2 | 9.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.8 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.2 | 1.7 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.2 | 1.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 2.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.2 | 2.0 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.1 | 1.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 2.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 3.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.9 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 3.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.4 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 0.3 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 8.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 4.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 15.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 5.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.2 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.6 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 12.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.2 | ST_WNT_CA2_CYCLIC_GMP_PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.5 | 18.8 | PID_ALK1_PATHWAY | ALK1 signaling events |
| 0.1 | 8.6 | PID_AJDISS_2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 10.6 | PID_BETA_CATENIN_NUC_PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 3.3 | PID_HDAC_CLASSII_PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.3 | ST_MYOCYTE_AD_PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 1.4 | PID_GLYPICAN_1PATHWAY | Glypican 1 network |
| 0.1 | 4.9 | PID_P53_REGULATION_PATHWAY | p53 pathway |
| 0.1 | 3.8 | PID_AR_TF_PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.6 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.0 | PID_ERBB4_PATHWAY | ErbB4 signaling events |
| 0.0 | 2.4 | NABA_ECM_AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 18.8 | REACTOME_GLYCOPROTEIN_HORMONES | Genes involved in Glycoprotein hormones |
| 0.5 | 8.6 | REACTOME_REGULATION_OF_INSULIN_LIKE_GROWTH_FACTOR_IGF_ACTIVITY_BY_INSULIN_LIKE_GROWTH_FACTOR_BINDING_PROTEINS_IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.3 | 6.7 | REACTOME_TRAFFICKING_OF_GLUR2_CONTAINING_AMPA_RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 2.7 | REACTOME_PURINE_RIBONUCLEOSIDE_MONOPHOSPHATE_BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 3.1 | REACTOME_CRMPS_IN_SEMA3A_SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 3.1 | REACTOME_SYNTHESIS_OF_VERY_LONG_CHAIN_FATTY_ACYL_COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 4.2 | REACTOME_MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.9 | REACTOME_BILE_SALT_AND_ORGANIC_ANION_SLC_TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME_PECAM1_INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 2.5 | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.2 | REACTOME_DARPP_32_EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.5 | REACTOME_SIGNALING_BY_ROBO_RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.7 | REACTOME_CIRCADIAN_REPRESSION_OF_EXPRESSION_BY_REV_ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.6 | REACTOME_METABOLISM_OF_VITAMINS_AND_COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME_SIGNALING_BY_FGFR1_FUSION_MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 1.4 | REACTOME_ASSOCIATION_OF_TRIC_CCT_WITH_TARGET_PROTEINS_DURING_BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.7 | REACTOME_FORMATION_OF_THE_TERNARY_COMPLEX_AND_SUBSEQUENTLY_THE_43S_COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME_SYNTHESIS_AND_INTERCONVERSION_OF_NUCLEOTIDE_DI_AND_TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME_TERMINATION_OF_O_GLYCAN_BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.6 | REACTOME_MITOTIC_PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME_OTHER_SEMAPHORIN_INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME_SIGNALING_BY_HIPPO | Genes involved in Signaling by Hippo |