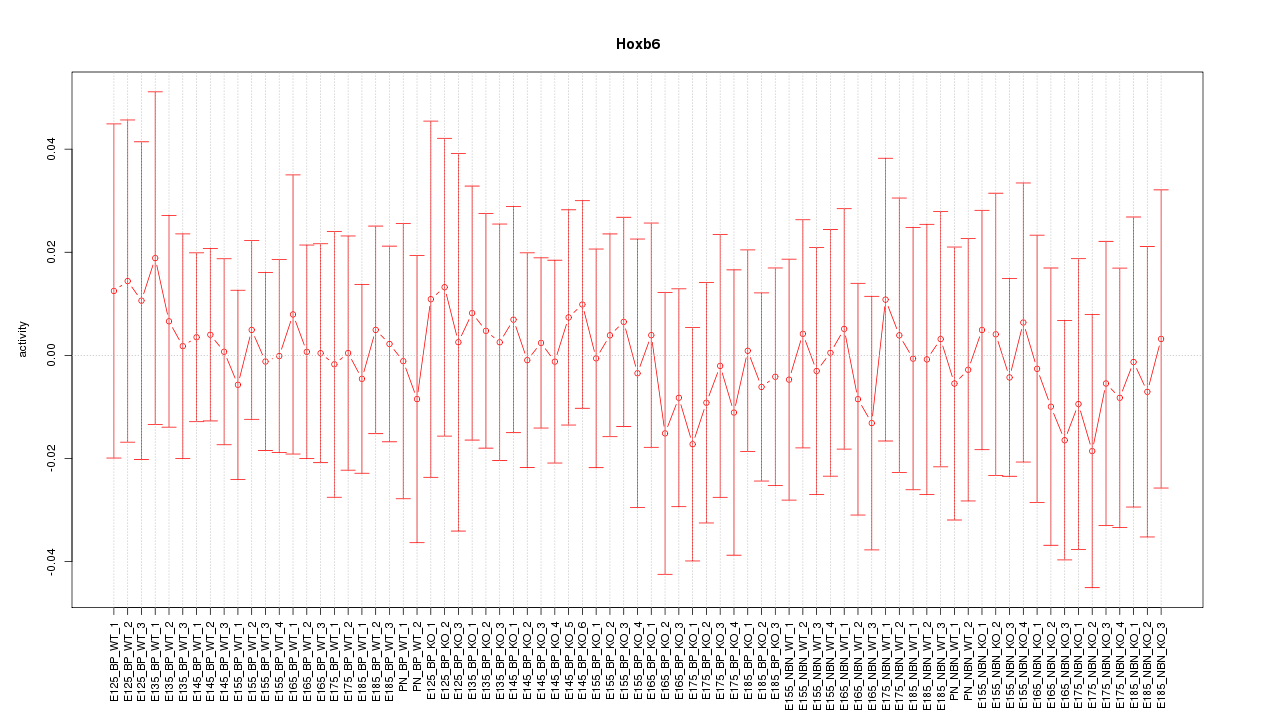
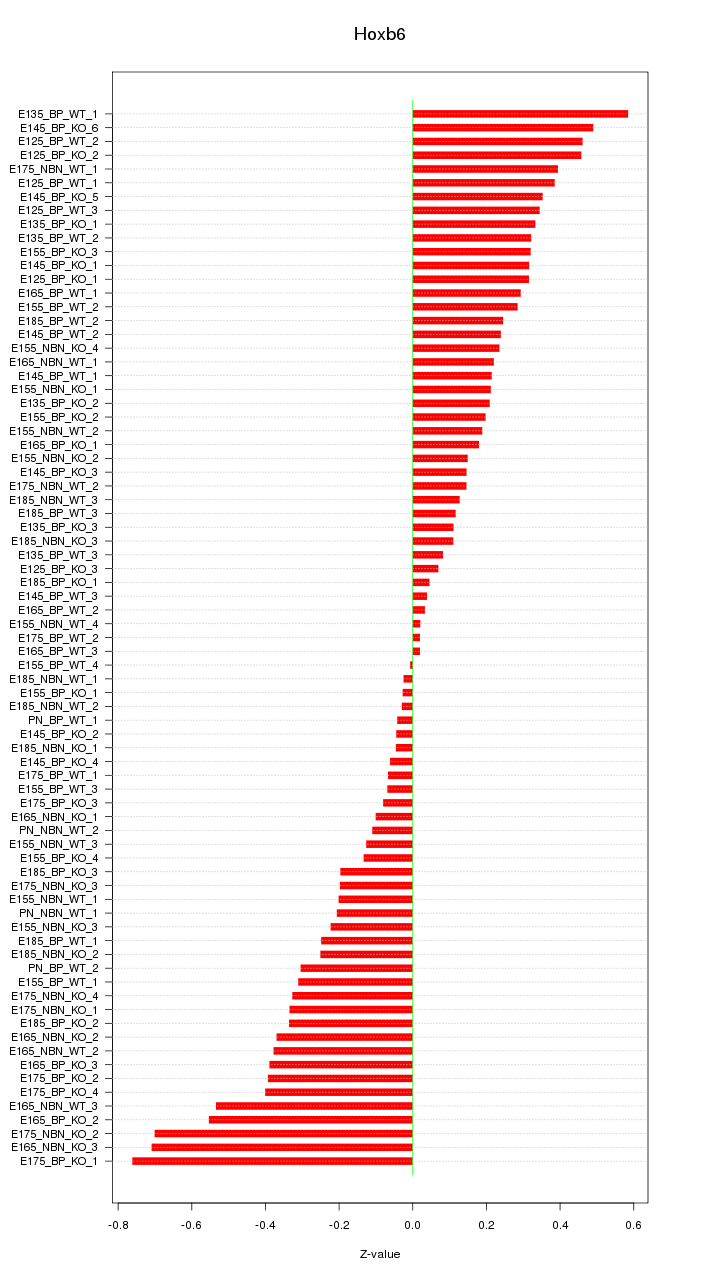
Motif ID: Hoxb6
Z-value: 0.295
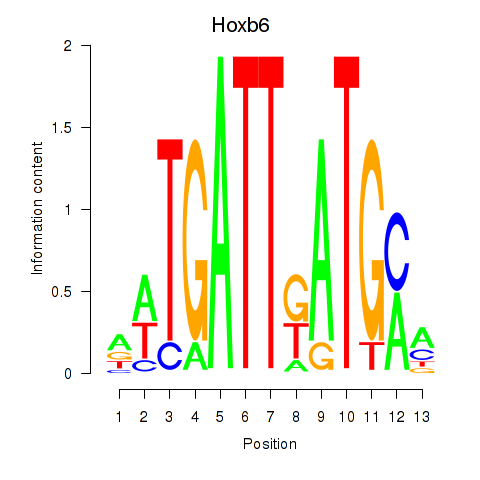
Transcription factors associated with Hoxb6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb6 | ENSMUSG00000000690.4 | Hoxb6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.5 | 2.5 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 1.1 | GO:0072235 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.2 | 0.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.7 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 1.6 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 1.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.2 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.7 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.9 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.7 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.3 | GO:0021559 | trigeminal nerve development(GO:0021559) |
| 0.0 | 0.4 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 1.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 1.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 1.0 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 0.7 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 2.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 0.7 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.1 | 2.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 4.6 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 1.1 | GO:0019842 | vitamin binding(GO:0019842) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID_LIS1_PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.3 | PID_CASPASE_PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME_CONVERSION_FROM_APC_C_CDC20_TO_APC_C_CDH1_IN_LATE_ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.3 | REACTOME_CASPASE_MEDIATED_CLEAVAGE_OF_CYTOSKELETAL_PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |